Introduction
This article is part of an ongoing series that highlights seqWell’s multimedia content offerings. The following is a transcript of our recent webinar: Enabling High-Throughput Biology with New Technologies for Synthetic Construct Sequencing.1
Joe Mellor, PhD, seqWell’s Co-Founder and Chief Scientific Officer, introduced the presentation with some background on the powerful enzyme that drives much of seqWell’s product line. 2 Then Henry Chan, PhD, Synthetic Biology Lead at Octant Bio, and Bryan Jiang, Research Associate in Synthetic Biology Technology at Octant, dove into the development of their upcoming high-throughput plasmid sequencing pipeline update, in which they leveraged the ExpressPlex™ 90-minute library prep workflow.3 The presentation closed with a brief Q&A segment.
The Versatility and Power of Tn5 Transposase
Joe Mellor: Welcome, everybody, to our webinar. You’ll hear today about how to enable high-throughput biology with new technologies for synthetic construct sequencing, which is an application webinar based on seqWell’s new ExpressPlex technology.
To tell you a little bit about seqWell, we really focus on how to solve some of the crucial bottlenecks in NGS workflows. Our goal is to help people sequence more samples on sequencing instruments more easily, and we do so with workflows and products that are designed to provide enhanced multiplexing, allowing you to take samples and prepare them into NGS libraries and get them onto sequencers more quickly, more easily with higher performance.4
A crucial technology that we use to build almost all of our products is an enzyme called Tn5 transposase, which is an incredibly versatile tool for scaling and simplifying NGS workflows.5 We have found Tn5 to be an incredibly powerful platform for creating library chemistry with several unique advantages, such as being able to normalize and create multiplexing with purpose-built reagents. These reagents form the core capabilities that our products are based on. We also have a lot of experience in creating products that support specific applications like, for example, plasmid sequencing, which you’ll hear about today.
We think of these as products of a workflow-engineering approach we have created that builds on the core reagents themselves and then creates streamlined products that deliver cutting-edge integrated workflow.
Many of these workflows are built into other products that seqWell has offered and continues to offer, starting with our plexWell technology and, most recently, culminating with our purePlex product launch last year and ExpressPlex, which you’ll hear about today. All of these products form the basis of a series of different technologies that support a range of different multiplexing needs across different applications such as genotyping, RNA-seq, genome sequencing, and plasmid and amplicon sequencing.
ExpressPlex is an exciting new product that we’re launching this year. This product is really the culmination of work on the part of our R&D team to create the fastest, most highly automatable library prep possible. Think of ExpressPlex as being the simplest possible way of turning DNA into a sequencer-ready library.
The way this works is we create plate-based products that allow you to put the DNA into a single reaction mixture and generate a high-performance library from hundreds or even thousands of samples quickly and easily. All of this can be done in a total time of 90 minutes to go from the DNA sample itself to a sequencer-ready library. As you’ll learn about today, this has several applications, including plasmid sequencing that our collaborators at Octant have been exploring.
With that, I’m going to turn this over to Henry and Bryan. They’re going to tell us today about their work in deploying ExpressPlex as part of the OCTOPUS protocol, which they have developed to support high-scale, high-throughput plasmid sequencing. Henry and Bryan, it’s all yours.6,7
Enhancing Drug Discovery
Henry Chan: Awesome. Thanks, Joe. We’re excited to be talking to you guys today about this joint collaboration between us and seqWell.
Real quick, I’m going to give a quick introduction on Octant. We are building a platform to navigate the complexity of biology and chemistry to develop the next generation of small molecules. We’re a therapeutics company, but more specifically today, I want to talk a little bit about our efforts with seqWell to help us develop and improve OCTOPUS. This is a high-throughput construct sequencing platform that we’ve used that really helps enable our drug discovery efforts.
Bryan Jiang: We’re excited to share with you all today on how seqWell’s ExpressPlex kit has enabled the possibility for the next-day turnaround of full plasmid sequence certification using Octant’s OCTOPUS v3 platform, which we’ll talk later about. We shaped this upcoming OCTOPUS update so that any interested lab or party can implement this within their own workflows. Henry is going to give some context for how onboarding ExpressPlex onto OCTOPUS has enhanced our company’s synthetic biology workflow and overall platform.
Henry Chan: Thanks, Bryan. Really quick, yeah, we are a drug discovery company. Our fundamental hypothesis is that the current state of drug discovery is poorly matched to address the multifactorial nature of human disease. We think that next-generation therapies are possible through understanding the right biological mechanisms that target the right receptors, the right pathways, and the right mutations.
A Real-World Example
Disease is complicated.8 We don’t believe in the silver-bullet hypothesis. What do I mean by this? Let me give you an example; Autosomal dominant retinitis pigmentosa is a disease that leads to progressive blindness. It is often caused by misfolding mutations in the rhodopsin gene. This leads to death of retinal cells.
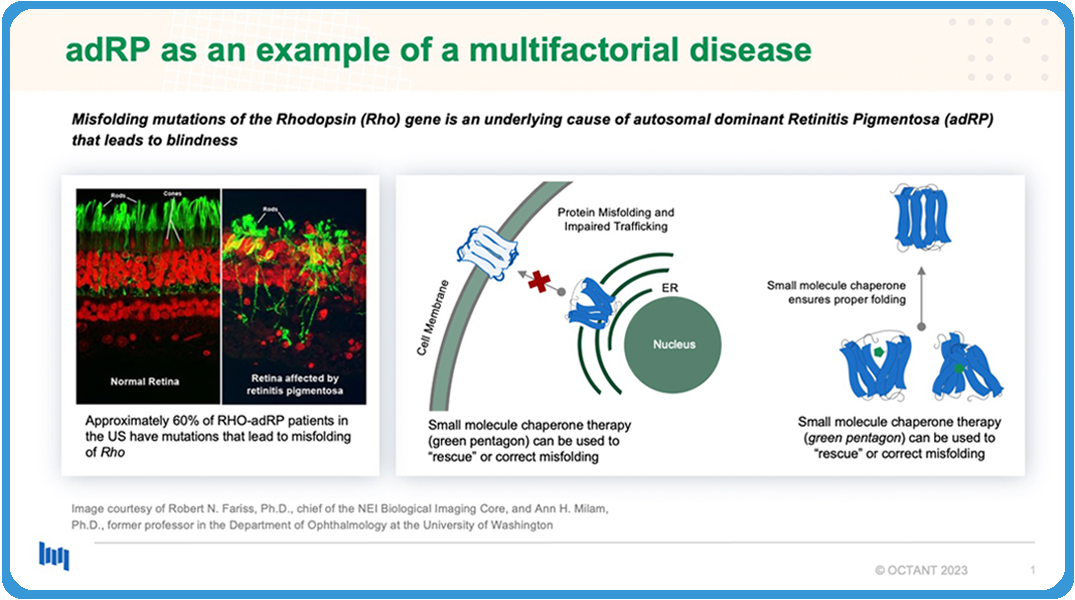
The mechanism behind how this is, if you look over on the figure in the middle, misfolding mutations of the rhodopsin protein will lead to mistrafficking of the protein from the ER where it is created to the plasma membrane. Small molecule chaperones could be a potential therapeutic for this. So you could imagine a molecule that might bind to the misfolded rhodopsin protein and then help it adopt a more native and wild-type confirmation. This adoption of the confirmation will rescue the trafficking and then prevent the cell death.
You might think one way to find molecules that could have the chaperoning activity is you might purify wild-type rhodopsin and then look for small molecule binders or stabilizers of the wild-type protein. That is certainly a fair way to address this problem, to screen for interesting compounds with therapeutic effect. However, we believe that, if you take an approach like this, you’re leaving a lot of potential mechanism off the table.
In the cell, a misfolded rhodopsin protein will undergo and experience all these different processes like biogenesis, translation from the ribosome, trafficking from the ER to the Golgi to the cell membrane.9 A potential therapeutic or chaperone will have many opportunities to interact in all of those complex stages of trafficking and folding. Additionally, there’s not just one single kind of mutation that leads to adRP. There are actually hundreds of known mutations.
A truly best-in-class therapeutic, you might want to address many or most of these mutations, and you don’t want to leave any mechanism off the table by only screening for binding to wild-type proteins. You want to look for things that affect the biogenesis and trafficking at the cellular level, and so this is one of our fundamental hypotheses.
The Driving Forces Behind Octant
It is around this hypothesis that we’ve built our drug discovery platform, which we’re calling The Navigator. To that end, we have two kinds of platforms. The biology platform, where we utilize multiplexed cellular assays, helps us optimize drugs and to do more precise drug design to multifactorial mechanisms and mutations. Then the other part of our platform, which I won’t talk about today, is our high-throughput chemistry platform. This allows us to design and iterate and generate tens of thousands of molecules every week. This rapid iteration allows us to really quickly optimize our drug design support rapidly.
We refer to our biology side as “cellular intelligence”, and this is based a lot on technology that our founder, Sri Kosuri, has pioneered. Sri did a lot of research as a professor in synthetic biology and genomic technologies, really pioneered multiplexed assays and these types of technologies like synthesis and gene editing.
So, a lot of our biology platform is built around our ability to engineer human cell lines with genetic barcodes. These barcodes are able to report on the activity of different targets in multiplex, and when we build these multiplex barcode assays, we use these as screens and assays for identifying and optimizing interesting therapeutic compounds.10
Two Main Multiplexing Methods
The two main ways in which our multiplexing technology breaks out are in the broad target scan (BTS) and in the deep mutational scan (DMS). In BTS, we can screen a hundred different targets or a hundred different mutations of the same target or a mixture of both. At this scale, we can do tens of thousands of molecules against hundred different targets or variants or mutations.
The other side of this is the deep mutational scan. Here, we can scan tens of thousands of mutations in parallel. So you can imagine we can make every single amino acid mutation to a given protein and then map out all of that at once in a big dish. So this could get us to understanding some SAR, or structure-activity functional relationships, at a very granular level. Whereas, on BTS, it gets us these broad chemical screens at many targets more broadly.
Multiplexed Assays With Synthetic Constructs
How do these play out for adRP? We built a multiplexed trafficking assay. As I told you before, rhodopsin has hundreds of known mutations that cause retinitis pigmentosa. We were able to build many of these and then multiplexed them into a single high-throughput screenable assay. From that, we were able to find compounds that actually rescued 80% of the trafficking mutations that we tested.
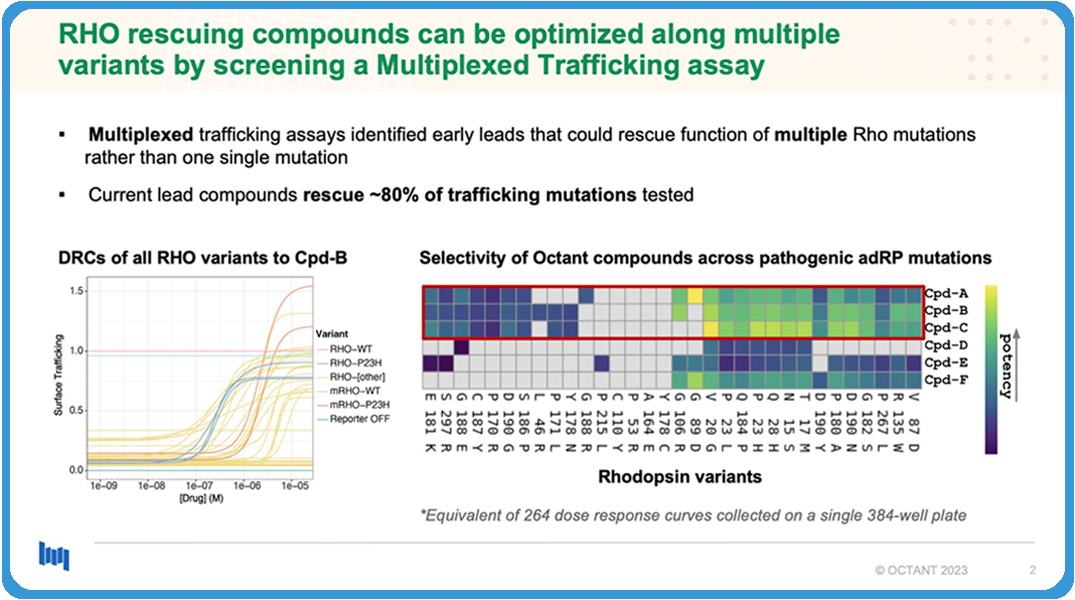
In this plot on the bottom right, you can see we’ve identified three compounds that have this level of rescue. Each square is actually a dose-response curve and, because we’re multiplexing, the density of data that we get is actually quite high. So we’re getting hundreds of dose response, equivalence of dose response curves that you might collect on, say, like a luciferase or fluorescence assay. However, this is against many variants at the same time.
Building and developing these assays requires a lot of work. Tons of cloning optimization were involved, and so, in building the assay that we just talked about, we optimized at least 14 different synthetic components to be able to achieve a signal that we were satisfied with.
In this process we tested at least 400 constructs. That is not the end or the only multiplexed trafficking assay that we want to build. There are many different mechanisms that we want to pursue.
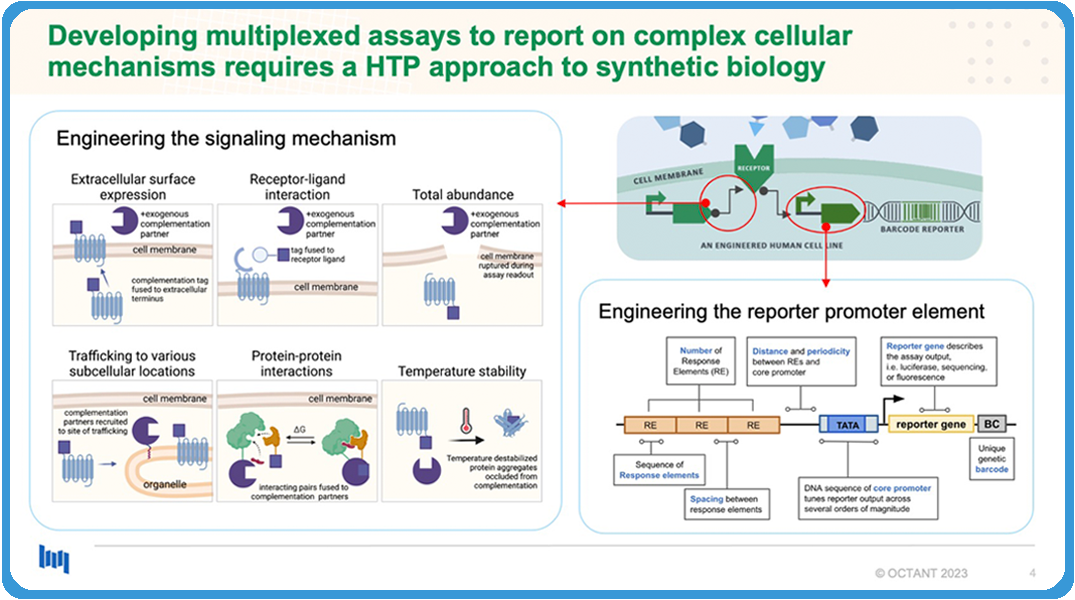
There’s an example. It’s not just some example, the ones on the left, that we can build in cell assays, and then there’s also the reporter element that expresses the barcode. So, we’ve also found a lot of things to optimize both on the mechanism side and the promoter side that actually expresses the barcode.
After building these multiplexed assays, we actually go through quite a lot of iterations of designs and cloning and cell engineering. So, rather quickly, we had to get really, really good at cloning, and so we adopted a lot of the latest gene synthesis.
Bottlenecks in Synthetic Construct Sequencing
We did take full advantage of a lot of the latest gene synthesis technologies and the pooled cloning approaches, but what we really ran into quickly was that something like plasmid sequence verification by Sanger became a huge bottleneck.11
We were sending in thousands of plasmid traces a week to a pretty large contract research organization (CRO). We incubated that. We were incubating out of UCLA. I won’t name who the CRO was, but we were spending quite a bit of money on them. They were throttling us at 90s, at one, two to four 96-well plates a day. This meant that we were having to wait a week to get our data back, not to mention the cost and then also just submitting tons of primers and analyzing thousands of traces.
This was not fun at all, and so we asked ourselves the question. We’re an NGS company. We live in the age of NGS. Why are we being bottle-necked by decades-old technology for DNA sequencing?
The reality is that this is a solved problem if your name is Ginkgo or Amyris and you’ve built your company around DNA-seq, around this sort of synthetic biology. We were a startup. We had two molecular biologists that were doing all of this cloning. We obviously couldn’t afford to build this world-class setup that they had.
Taking Innovation Into Their Own Hands
This is back in 2019 when we developed OCTOPUS, which is a cheap and scalable high-throughput full-plasmid sequencing platform that is super scalable and direct from colony. So, no mini preps required. It’s accessible, no fancy equipment. You only need a MiSeq and maybe some multi-pipettes. Fast turnaround is comparable to Sanger. Automated analysis report, you don’t have to sit in front and stare at traces. It just spits out an Excel file. It’s cheap. It costs the same if you bought a Sanger.
Like any good automation and process, it changed the way that we approached experimentation and the things that we were able to do on the bench side, in the molecular biology lab. We were able to just take riskier and riskier cloning approaches and go crazy with what we wanted because we had this thing on the backend that would just tell us what we were doing at a high scale, and then the other thing that we thought was we should open source this. So you could actually find our protocols for exactly how to do this on GitHub. We wrote a blog post about it.12
In a nutshell, really quick, how this process works: On day one, you pick colonies. Day two, get in really early in the morning. You lyse the cells. You extract the DNA in bulk at the plate level. Then you do the library prep. At the time, we were using the iGenomX Riptide kit for library prep. That takes about six hours, and then, by that evening, you could load your MiSeq or your next-gen sequencer. Then, less than 48 hours later, you get your results, your analyzed pipeline results.
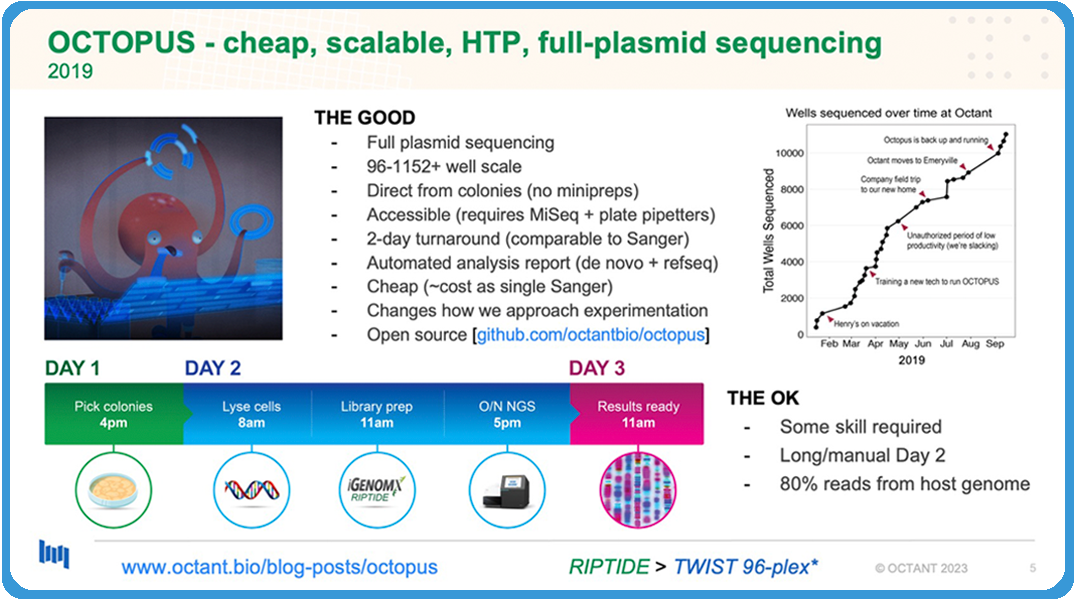
As you can see on the plot over on the top, in the first six months that we developed this, we loved it. We basically sequenced 12,000 plasmids on this using OCTOPUS. Again, as I told you, we only had two molecular biologists. We had one RA that was running this, but there were still some things that were not perfect about it, and I’ll pass it off to Bryan to tell you about it.
Optimizing Synthetic Construct Sequencing
Bryan Jiang: Yeah. Thanks, Henry. Now, I wasn’t around the time to run v1 of OCTOPUS, but from what I understand, running day-two sample prep and library prep by hand was quite a pain in the butt.13 Also, the cell lysing method being used resulted in higher gDNA bias than we would have liked. Reducing the hands-on labor required and the gDNA bias culminated in the v2 update of OCTOPUS.
One major change was the onboarding of rolling-circle amplification (RCA) to replace the older cell lysis sample prep method that we had.14 Modifying RCA with the addition of plasmid-specific primers greatly improved the quality of our sequencing by reducing the amount of gDNA contamination.
Additionally, we utilized the Opentron’s OT-2 liquid handling robot. This robot is a cheap automation that further reduced the amount of hands-on, laborious work required to get through the day-two library prep. While we addressed the gDNA and hands-on labor problems with the v2 updates, the library prep process itself, while still using the library kit that we had, still had a lot of moving parts that made the process more complicated than we would have liked.
Henry Chan: Yeah, and so then this is actually drawn from last year. This is when we got in contact with the folks over at seqWell, Roger and Joe. They found us through our blog post, and they reached out to us and said, “Hey, we have this kit called ExpressPlex we think you might be interested in”. They showed us this protocol, and it doesn’t get simpler than that.
Saving on Time, Cost, and Training
ExpressPlex takes 90 minutes, to compare that to the Riptide protocol which, if you guys know, Riptide was bought out by Twist. It’s now called Twist-96 Plex, but we refer to it as Riptide because we actually never bought any kits from Twist. We had so many other Riptide kits. Anyway, just compare that to the Riptide protocol, which is super long, takes about five to six hours, has many steps, many ways that you can mess this up. There was always training involved. So we saw this ExpressPlex protocol, and we were like, “Yes, we would love to be the alpha testers for this product”. So we brought it on.
In our early prototyping, our early testing, we found out that the sequencing bias between Riptide and ExpressPlex was basically the same in terms of performance. If not, maybe ExpressPlex even has a slight edge on it.
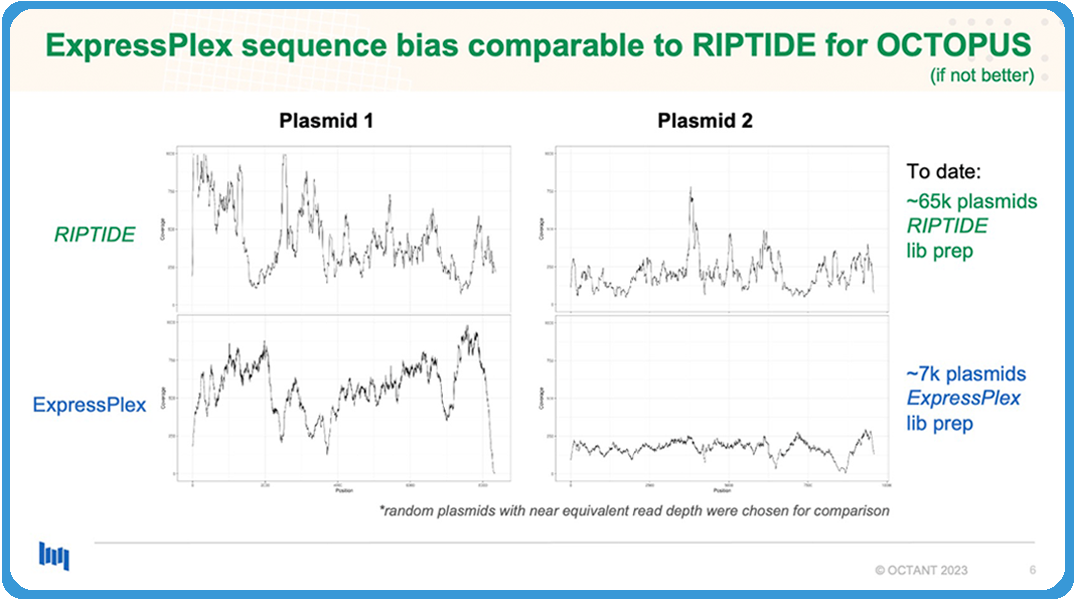
What I’m showing you here is just bias plots from two random plasmids that we’ve done. We’ve done more rigorous statistical analysis on the backend to convince ourselves, but I think the takeaway is, to date, we’ve sequenced now 65,000 plasmids using the Riptide library kit, prep kit and never experienced any issues. Obviously, bias is going to be introduced whenever you have library prep, but it was fine for us, and so, when we saw something that was almost basically just as good, if not better, we were super happy with that.
As of today, we sequenced around 7,000 to 10,000 plasmids using ExpressPlex library prep kit and, again, no issues there. With that, we were really excited. Actually, Bryan was really excited to bring ExpressPlex on board.
Bryan Jiang: Yeah. Basically, with the simplified and faster library prep that ExpressPlex allowed, it made my life a lot easier, and running OCTOPUS has shortened the library prep day quite a lot. This also got us thinking that, if we were able to make the library prep day this much shorter and able to turn RCA that was formerly an overnight process and squeeze it into the same date as the library prep, we might be able to get next-day turnaround of our plasmid sequencing.
Indeed, we’ll talk about this later, but this culminated with our upcoming v3 update of OCTOPUS. By combining the speed and simplicity of ExpressPlex with a shorter RCA protocol, we were able to achieve next-day turnaround of full plasmid verified sequences. Additionally, the library prep processes still can be assisted by the OT-2.
Henry Chan: Yeah. I mean, this is crazy, right? It’s literally faster than Sanger. As Bryan mentioned, we’re going to be releasing these protocols soon. Just be on the watch-out for that. We haven’t actually written everything up yet, but it works. It’s awesome. Again, this changes everything for us. Basically taking a two-day process to one day just dramatically shortens time to data.
A Workflow Change With Massive Payoff
In our workflows, you can imagine if you order, say you’re super excited, from thinking about your problem on the weekend, you come in on Monday fired up, you’re ordering primers, you clone them, and then AVITI delivers them on Tuesday because they’re amazing, and then you clone on Tuesday, and then you do OCTOPUS on Wednesday. You get your results by Thursday, and you can then now set up your experiment or, if you messed up, reorder primers on Thursday.
If you set up your experiment on Thursday then, for us, we transfect on Friday. That allows you to take advantage of the weekend incubation or re-clone on Friday. Because of the weekend and the way that the week breaks out and lab work just happens, this one-day shortening of the processes gives us two turns of the crank on any given experiment rather than one. This is huge.
The other side of this, too, is this easier workflow. Until now, everybody has had to rely on Bryan to run OCTOPUS for them. Because of this super long and involved library prep, only Bryan knows how to make the OT-2 sing like it does.
However, now it’s just one-step transfer and then put it onto the library. We can actually democratize this within Octant, and people can run it. Our vision is that anybody who does molecular biology can just run OCTOPUS whenever they need to and not have to wait for Bryan.
The other side of this, too, is a lot of people reached out to us about OCTOPUS, and I know of at least 10 companies that are probably using OCTOPUS right now. I would imagine many more would use it if they knew it was this easy and was literally faster than Sanger. We could also imagine there are more turns of the crank, but also more turns of the crank democratized out to everyone within Octant, but also outside of Octant, too. So we’re really excited to be able to share this protocol.
Summary
In summary, Octant is a drug discovery company. We’re trying to build the next generation of small molecular therapeutics by leveraging synthetic biology and multiplexed cell-based assays and our high-throughput chemistry platform. OCTOPUS, this tool that we’ve developed, has been greatly enabling for the synthetic biology efforts here in the engineering of cell-based assays.
With this collaboration with seqWell and the advent of ExpressPlex into the workflow, this reduction in turnaround time, labor, and expertise for OCTOPUS has greatly increased the usefulness and accessibility of this.
Bryan Jiang: Yeah, and, with the OCTOPUS input, you can have transformed colonies on day one to mini-prepped, full plasmid-verified culture by the next day. Our platform will be freely available for open source collaboration and mutual benefit across all labs as well as in OT-2 protocol to further simplify the one-lab process.
Henry Chan: Yeah, and with that, I’d like to give some shoutouts here. Nathan Lubock, he was a scientist, a bioinformatician at the time, and he wrote most of the analysis pipeline for OCTOPUS.
Johnny was that first RA we talked about, and he made a lot of improvements to v1 and v2.
Liz was a second RA runner, and she made some improvements that will spruce up the analysis pipeline for us a little bit. And finally, Bryan has done a lot of improvements to v2 and v3, and he is the current runner for us right now. For the rhodopsin projects, Eric Jones is the lead scientist. Our two founders are Sri Kosuri and Ramsey Homsany.
Then the folks over at seqWell, Joe, Roger, Curtis, Jack, Kellie, Samantha: You guys have all been great and super helpful in this collaboration. We’re excited to continue working with them. With that, thank you guys for your attention and give the floor to any questions.
A Brief Q&A
Are the protocols for OCTOPUS and ExpressPlex published or available to review?
Henry Chan: In terms of OCTOPUS, we are currently trying to write up the protocols and formalize them. We hope to have that finished by the end of March. To get a sense for what the workflow is for OCTOPUS sans the fast RCA and ExpressPlex, you can take a look at our GitHub and our blog posts. Here is a much longer version of the protocol.
Joe Mellor: Yeah, and just to add to that on the ExpressPlex information, certainly, anybody who’s interested in learning more can reach out to any of us at seqWell. This product will be officially launched in April 2023. We’re certainly looking forward to publishing a lot more information on protocols and et cetera on our website soon.
How much time did it take you to integrate ExpressPlex into your overall workflow?
Bryan Jiang: Yeah, so the ExpressPlex kit is as simple as it can really be, just stamping and a single thermocycling reaction. For us, we were able to implement ExpressPlex within OCTOPUS on the first attempt. In the first run, we were able to get sequence verified results. The kit does its job. It converts plasmid input into a Illumina sequenceable library and the OCTOPUS process basically. Yeah, that’s just what it requires. It’s just a faster, more simplified version of the system we had before.
What is the maximum number of samples of small plasmid, say, 10 KB, that can be multiplexed within a single flow cell?
Henry Chan: I think it depends on the sequencing kit you use. We routinely use the v2 micro from the MiSeq by Illumina. I think that gets you 4 million paired in 150 reads, and so very, very comfortably that is eight 96-well plates. I’m pretty sure you could push that up to twice that at the 10 KB size, but I think the limit there for us is actually the number of unique indexes.15
In your experience, how long does it take to prepare the sample normalization to start with the protocol, and can you please briefly explain how you use RCA?
Joe Mellor: I’ll take the first part just to address the normalization question. I think one of the really attractive features of ExpressPlex is it does normalize over a pretty wide range of input DNA amounts. Again, I think a typical use case certainly could be a global dilution, not necessarily individual normalization of samples. Maybe Bryan and Henry can speak a little bit to how the output of the RCA process fits with that concept briefly.
Bryan Jiang: Yeah. The cool thing about the RCA process is, when RCA has run the saturation, that also is a step of normalization of all the input samples. On top of that, it doesn’t require spot quanting at the beginning. After you have your sample and RCA has run the saturation, you can dilute that by a global factor, as you can expect, all wells to have roughly the same amount of input, and then that can be stamped into the ExpressPlex kit.
Can ExpressPlex be used for applications other than plasmid sequencing?
Joe Mellor: The answer is absolutely yes. I think certainly, initially, our focus applications will be a wide variety of synthetic constructs, including things like amplicons. These are the key applications, and we see this being a great fit for now. We will certainly look forward to expanding beyond that. I can tell you, certainly, we’ve had some really positive data on other diverse applications such as RNA-seq that we’re looking forward to telling the world about more quite soon.16
Again, I think the answer here is that it’s a new product, and we’re excited to get it out there and certainly learn from our customers about where it can go. I think, initially, certainly applications like the ones you see here today are going to be the ones that this is really well-suited for.
How many samples are you processing at one time or sequencing in a batch, and how many reads are required per sample?
Bryan Jiang: So with the Illumina micro kit and sequencing 150-based paired end, we comfortably sequenced with just eight indexes, eight times 96 samples, around 800 samples, comfortably getting an average of around 5,000 reads per sample. That’s plenty of coverage for a 10 KB plasmid input. With more indexes, it’s possible to imagine running maybe up to 1200 samples or 12 indexes multiplied by 96 samples within a micro kit.
The OCTOPUS platform is able to accommodate those levels of read coverage because it doesn’t rely entirely on de novo sequencing, which would require a lot of reads, but it has an intermediate step basically allowing for a full-plasmid sequencing with the low input sequencing kit.
Henry Chan: Yeah, and then I think, one day, if there is a need to do more than eight 96-well plates per run, I think, one, that’s totally possible with the micro kit. Two, the v3 kits are like 30 million reads, and so you can imagine 10 times the number of samples if you are one of these big cores, or if you want to eat the lunch of these Sanger companies and try to conglomerate the entire country’s worth of plasmid sequencing all a single location, you’d probably do that too. I think, again, you’re just bottlenecked by the number of unique indexes.
Bryan Jiang: Yeah, and alternatively, if those are pretty large scales, you can also easily imagine that using a nano kit would be sufficient with just one or two 96-well plates with the samples. That would probably also give you 5,000-ish reads per sample, and that’s plenty of coverage for 10 KB plasmid for OCTOPUS.
Do you see this technology and approach replacing Sanger sequencing at workflow and costs that labs can change, especially core labs and high-volume Sanger CRO’s service providers? Also, what would you pass on as words of wisdom from your experience to folks thinking about this change?
Henry Chan: I think doing a complete 100% replacement of Sanger is probably not what this would do, but, at the same time, probably close, for the vast majority of plasmid sequencing, I don’t see why that wouldn’t be the case. We still have an account over at our CRO, but I would say 95% of the plasmids, or more even, that we generate here at Octant are sequenced by OCTOPUS. Especially with the easier workflow and the shorter turnaround time, that is actually faster than what Sanger can actually provide. There’s no reason why, for most things that OCTOPUS can handle, you wouldn’t do this.
If you don’t have high volume, I think it’s not as worth it at the sub-96-well level. I think, right around there, it’s really worth it to do, but if you are a high-volume lab or you’re a core facility at a university or something like that, it just doesn’t make sense to me that you would ever use Sanger.
I think there are some things that, one, we’re not selling OCTOPUS. We try to develop it to the point that’s useful to us and share it with the world, but there’s probably some tweaks of the analysis pipeline that might make some things a little bit tighter. Then the other thing is, of course, with any kind of sequencing library prep, there will be bias.
If you have highly, highly repetitive sequences littered throughout your plasmid, which there are all sorts of problems with that just from the cloning and the design side anyway, I think we’ve seen that OCTOPUS can choke on that a little bit. We can still sequence it, but it’s not perfect, so every now and then we’ll just use Sanger to spot check some things that seem crazy.
Then in terms of words of wisdom, just email us. Honestly, if it wasn’t this easy, if it was still v2 or v1, I would have a lot more things to say. When we spin up the protocols, definitely email us to see what might help. It’s so easy. I don’t anticipate any wisdom needing to be shared other than having to have this scale.
Bryan Jiang: Yeah, pretty much it. Once you’re approaching a scale of 96 samples or more, that need to be fully sequenced, or it doesn’t have to be fully-sequenced even. It’s because of how easy it is. If you’re on the 90, at that point, analyzing Sanger traces becomes the pain, and having OCTOPUS as an option, even as a first-pass option, you pretty much get everything you need. If you’re still more comfortable with Sanger, you can use Sanger. But, at least for us, we quickly realized that OCTOPUS is pretty sufficient for almost all use cases, especially for cases where you have complex plasmids cloned in de novo or just having a lot of samples.
Because all the input samples don’t have to be mini-prepped, you literally start from bacterial colonies. That further simplifies the process. You don’t have to consider like, oh, I have to mini-prep eight of this particular construct. You can just pick your colonies, put it in, and then just mini-prep the single perfect at the end. That’s just another benefit to the OCTOPUS process when you’re approaching the 96-well scale.
Are you looking at any of the new NGS platforms?
Joe Mellor: Yeah. I think the short answer there is, certainly from our point of view, multiplexing is a key need that spans various sequencing platforms. Certainly, as newer platforms have come out especially in the last year, I think we’ve seen that now become more and more apparent.
At seqWell, we’ve done some really good work that I’m happy to direct you to our website and check out. We’ve shown really good performance with some of our other kits with the Element platform, the AVITI sequencer.17, 18 But certainly, looking across the spectrum of other platforms, I think there’s some really exciting stuff going on certainly in the long-read arena.19 Some of the new platforms in that space I think offer throughput and cost that now really is going to motivate a high level of multiplexing as well.
We’re very excited to start to think through how to implement some of these technologies on other sequencing platforms as they get out there. Yeah, more to come, and I certainly invite you to take a look at, again, some of the work we’ve already put on our website in that regard.
References
- https://fast.wistia.net
- https://www.youtube.com
- https://seqwell.com/expressplex-poster/
- https://seqwell.com/economizing-ngs-library-prep-with-plexwell-technology/
- https://seqwell.com/tn5-transposase-a-breakthrough-enzyme-for-dna-library-preparation/
- https://www.octant.bio/blog-posts/octopus
- https://seqwell.com/the-need-for-high-throughput-ngs-based-plasmid-sequencing/
- https://seqwell.com/outsmarting-chronic-diseases-a-case-study-in-how-ngs-accelerates-microbiome-research/
- https://rupress.org/jcb/article/215/6/769/46099/Protein-sorting-at-the-ER-Golgi-interfaceProtein
- https://seqwell.com/getting-a-read-on-ngs-barcodes/
- https://seqwell.com/sanger-method-the-mp3-player-of-sequencing/
- https://www.octant.bio/blog-posts/octopus-v2-0
- https://www.octant.bio/blog-posts/octopus
- https://seqwell.com/streamline-plasmid-prep-for-ngs-with-rolling-circle-amplification/
- https://seqwell.com/when-unique-dual-indexes-matter/
- https://seqwell.com/increasing-accessibility-of-full-transcript-scrna-seq-through-continued-innovation/
- https://seqwell.com/wp-content/uploads/2023/02/SW2023_purePlex-AVITI-AN-02062023v2.pdf
- https://seqwell.com/wp-content/uploads/2022/06/TechNote_Element-AVITI_plexwell.pdf
- https://seqwell.com/wp-content/uploads/2023/02/SW2023_TJMS-AGBT_NOQR-8.5×11-02032023.pdf
