
purePlex™ DNA Library Prep Kit
Speed, Performance, and Auto-Normalization with Unique Dual Indexes
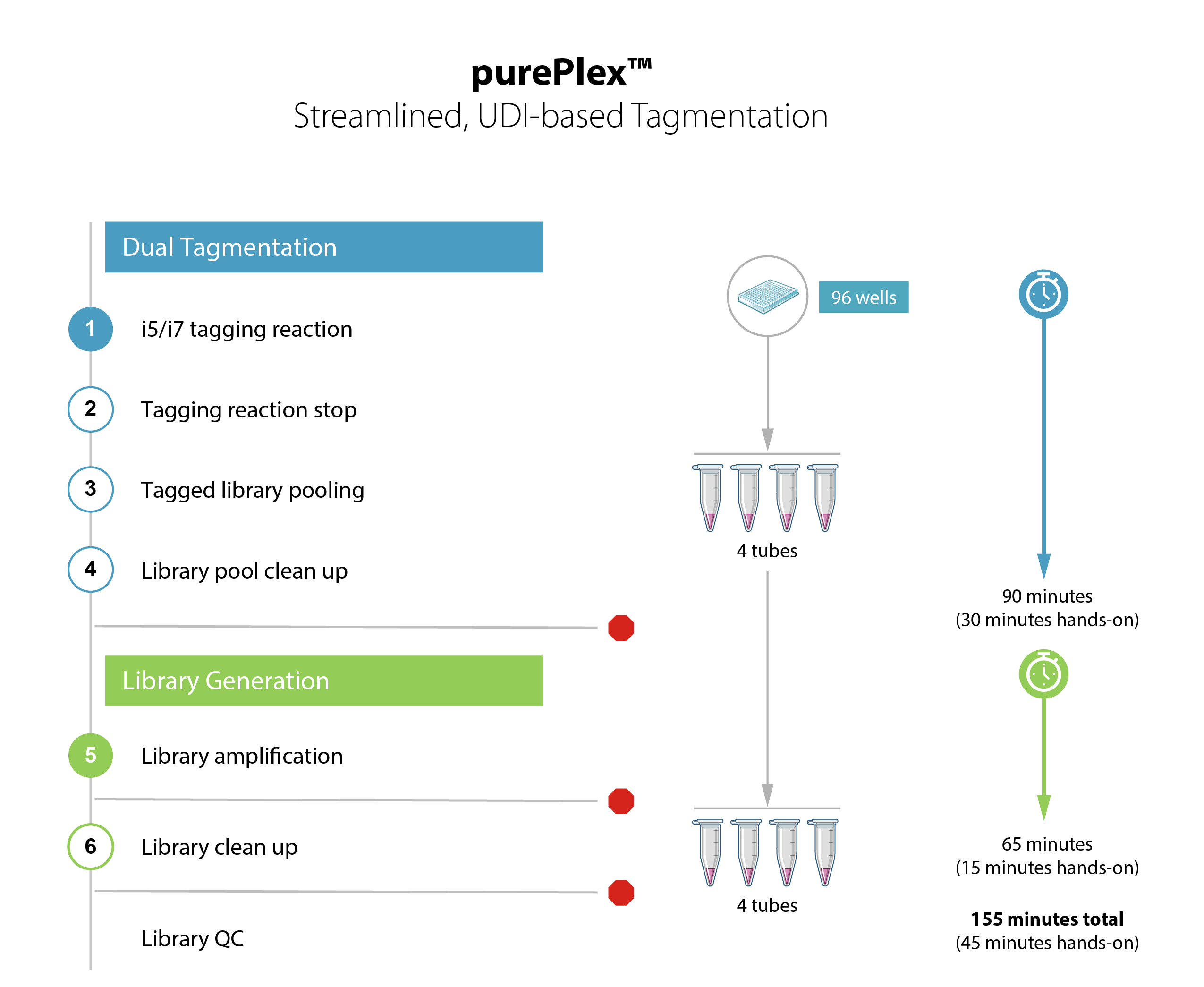
- 2.5-hour workflow for 96 samples, 45 min. hands-on time
- Auto-normalization of read counts and insert size over 10-fold input range
- Unique dual indices included
A benefit to purePlex is that, because of its simplicity, users can quickly and seamlessly incorporate the kit with existing methods for high-throughput pipetting.
Key Features
- Fast, flexible workflow with no requirement for full plate processing
- Auto-normalization reduces QC burden, improves data consistency
- Early pooling for easier sample handling
- Reduced GC bias compared to other transposase-based methods
- Significant cost and plastics savings
Check out what our customers are saying:

“I’ve always used i5 and i7 barcodes. Other library prep methods incorporated a pool barcode so only one was unique. With purePlex the i5 and i7 are unique to every single sample so I can catch barcode slippage much more easily. It makes me feel a lot more comfortable. purePlex is also a lot quicker than the Nextera kits we’ve used in the past. Now we use a lot less tips and can go from DNA to having our library in a small tube by the end of the day then having it sequenced the next day. It’s amazing.”
– Sophie Nozick

“The sample auto-normalization with purePlex simplifies our sample balancing process without seeing evidence of substantial bottlenecking of unique reads or an overabundance of PCR duplicates. That’s very good for us because it means we are not wasting sequencing data on duplicate information or on imbalanced samples. We found that, in general, purePlex was either comparable or performed better than other methods for both balance and low duplication.”
– Ray Watson
POSTER: Haplotype Reconstruction Using Low-pass Whole-genome Sequencing in Genetically Diverse Mouse Populations
POSTER: The Long and Short of Sequencing for Microbial Profiling of Metagenomic Samples

POSTER: Automated Low-Pass Whole-Genome Sequencing to Scale and Accelerate Genotype Imputation
>> View Now

Product Highlights
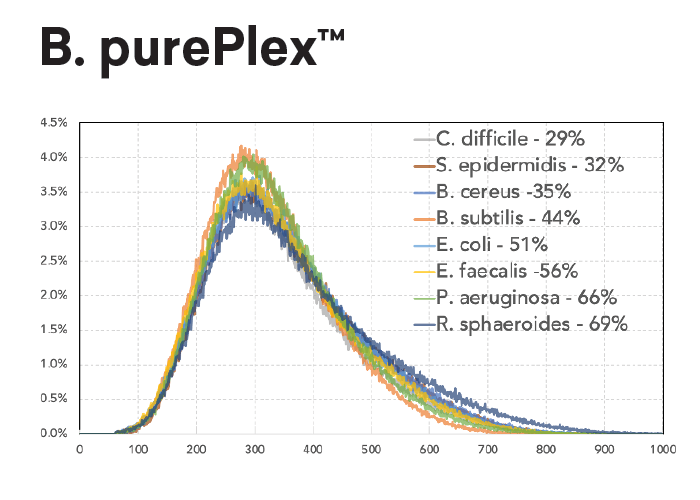
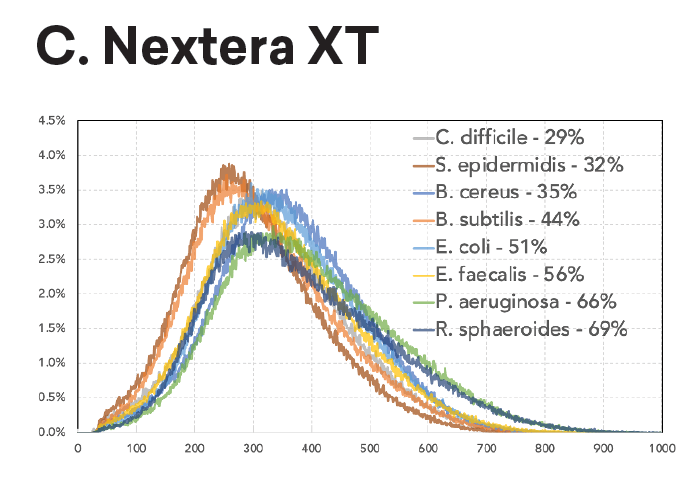
Auto-Normalization of Insert Size and Read Depth:
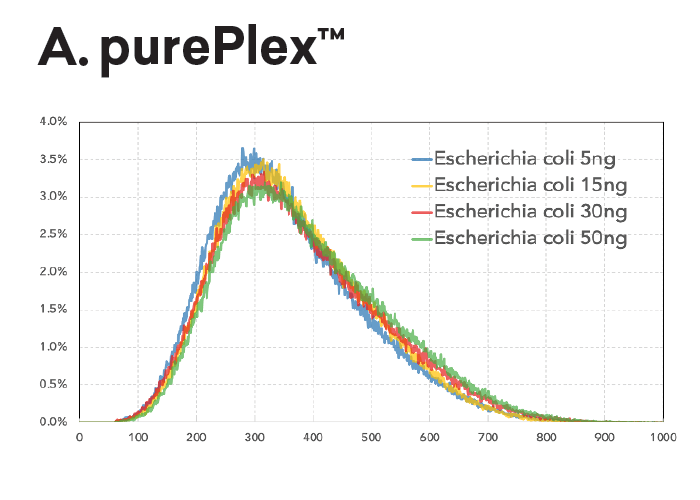
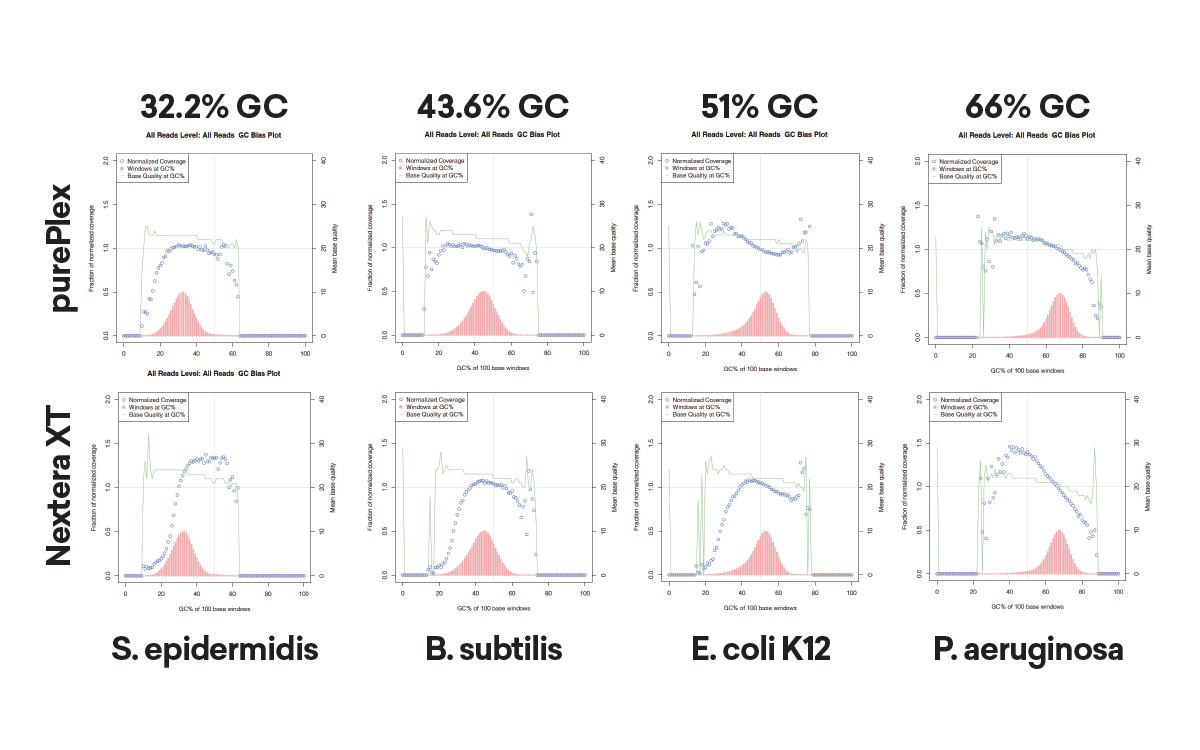
The insert size within a pool of samples is consistent regardless of input (panel A, left) or GC content (panel B, lower left). In contrast, Nextera XT libraries have varied fragment distributions from GC content even after normalizing the sample input.
Auto-normalization of Read Depth
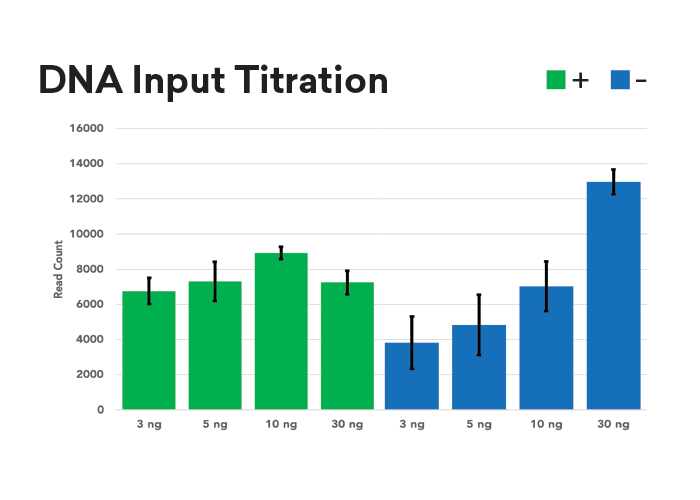
Samples were normalized to inputs of 3, 5, 10, and 30 ng then underwent purePlex library prep with (+) and without (–) normalization reagent. Read counts for each sample are equal, regardless of input, when normalization reagent is used. In contrast, without normalization reagent, sample read count scales with input.
Specifications
| Specs | Description |
|---|---|
| Sample Type | Amplicons, Plasmids, Genomic DNA, cDNA |
| DNA Input Range | 5 – 50 ng |
| Number of Unique Index Combinations | 384 |
| Supported Paired Reads (Clusters)/Sample | ≤ 20 million |
| Output Fragment Range | 400 – 1,200 bp |
| Applications | Synthetic construct sequencing (amplicons, plasmids, etc.), Low-pass whole genome sequencing, Whole small genome sequencing (<50 Mb), scRNA-seq, Metagenomics/Microbiome screening |
| Reactions per Kit | 96 |
purePlex DNA Library Prep Kit Includes:
- i7 Tagging Reagent Plate
- i5 Tagging Reagent Plate
- Coding Buffer (3X)
- X Solution
- MAGwise Paramagnetic Beads
- Normalization Reagent
- Library Primer Mix

