Introduction
Single-cell sequencing is a rapidly evolving field that is providing researchers with unprecedented insights into the function of individual cells. Using optimized next-generation sequencing technologies, this method allows for a more detailed analysis of cellular differences and provides a better understanding of the function of an individual cell in the context of its microenvironment.1
In recent years, single-cell RNA sequencing (scRNA-seq) has emerged as a powerful tool for understanding transcriptomic differences across cell types to address myriad questions in cell and molecular biology. In fact, this technique has revolutionized our ability to study complex biological systems and has led to new insights into developmental processes and diseases, including cancer.
Challenges and Considerations
Automated scRNA-seq technologies using droplet-based (e.g., 10x Genomics) or microwell (e.g., BD Biosciences) partitioning enable the analysis of many cells in parallel, but generally only provide information on the 3’ or 5’ portion of mRNA transcripts.
Methods that capture scRNA-seq from the entire mRNA molecules can be difficult and demanding. These methods typically require more processing steps and hands-on time, making them more expensive and less scalable to large numbers of cells. The complexity of these protocols also limits compatibility with standard liquid handling instruments, confounding automation that could enable robust and reproducible analysis of more cells. In a previous blog, we discussed the important yet challenging goal of obtaining full-length scRNA-seq data at the single-cell level.
To recap briefly, full-length scRNA-seq must be extremely sensitive to very small quantities of RNA—often as little as 1-2 picograms—to capture as much transcriptomic diversity as possible. In addition, the process for converting amplified cDNA into an NGS library is difficult to perform at scale, especially if time-consuming QC, purification, and normalization steps are required to ensure robust performance.
An Innovative Way to Amplify and Sequence
Our goal has been to improve both the scalability and efficiency of different NGS methods. The plexWell Rapid Single Cell kit couples our expertise in multiplexed NGS library prep with a best-in-class single-cell synthesis and amplification workflow.
While the plexWell Rapid Single Cell kit offers compelling performance in terms of transcript sensitivity and uniformity per cell for plate-based scRNA-seq projects, we are continuing to innovate toward streamlined, cost-effective workflows that can interface with common laboratory automation and enable higher throughput full-transcript data acquisition.
The Potential for Future Breakthroughs
In a recent presentation, seqWell shared a novel reagent system called ExpressPlex™ that simplifies and expedites library prep. It requires only two pipetting actions and a single thermocycling procedure to prepare amplified libraries from cDNA. The method was used downstream of a modified FLASH-seq cDNA synthesis protocol to enable faster, easier, and automation-friendly full transcript scRNA-seq.
Pairing the plexWell Single Cell Rapid cDNA synthesis kit with the ExpressPlex Library Prep Module allows for rapid library prep over a range of RNA and cDNA inputs without sacrificing sequencing data quality or content. In addition, both cDNA synthesis and library prep modules can be automated using common liquid handling capabilities, saving both hands on and total turnaround time, and limiting the number of consumables needed.
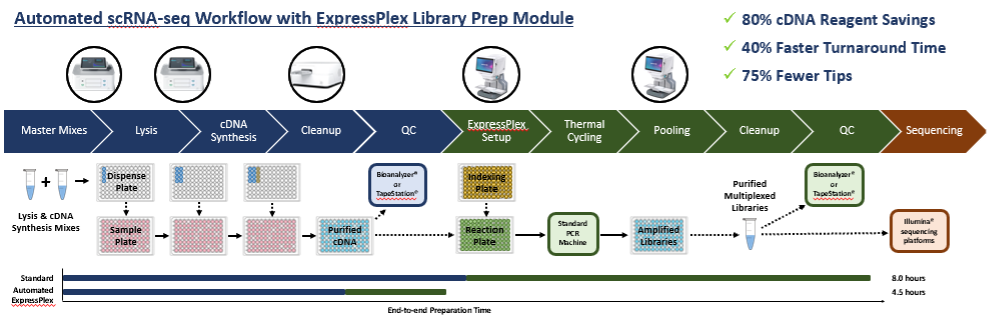
We recently road tested this combined solution in a head-to-head comparison with a manual, plate-based scRNA-seq method. cDNA synthesis was carried out using the plexWell Single Cell Rapid workflow or a miniaturized, automated workflow.
Downstream library preparation was performed using either a standard or semi-automated ExpressPlex solution. The automated method yielded high-quality sequencing data with exceptional library complexity, which was shown to be consistent with the established, manual method. And because the cDNA synthesis reaction can be miniaturized using common liquid handling automation, the cost per sample can be significantly reduced. Importantly, this type of automation is in scope for labs using benchtop liquid handlers as well as larger facilities that require higher throughput solutions, as the volumes and formats are compatible with either type of solution.
Recent work discussing FLASH-seq, has also been shown that miniaturization can increase the number of genes detected and the diversity of gene targets.2 This leads us to believe that a miniaturized system may also allow for the extraction of more data per cell.
Conclusion
Single-cell RNA sequencing is a powerful tool that can yield insights into a wide range of biological processes. However, the complexities of the technique can often be daunting, and the library preparation process can be time-consuming. The plexWell Single Cell Rapid cDNA synthesis module addresses these challenges by providing a simple and efficient way to generate high-quality cDNA without sacrificing sequencing data quality or content.
The ExpressPlex library prep module further streamlines the library preparation process, making it easier than ever to obtain high-quality RNA-seq data. Together, these two modules allow users to efficiently generate full-transcript RNA-seq data with minimal hands-on time, making this powerful technology more accessible than ever before.

