Introduction
Few people have had as great of an impact on the field of genetics as Frederick Sanger. A two-time Nobel Prize winner, his groundbreaking work in DNA sequencing changed much of what we know about genetics and how genes function. He developed what is now known as the “Sanger method,” which uses chain termination to sequence labeled DNA fragments.1 Much like what the MP3 player did for the music industry, the Sanger method propelled genetics toward the innovations we see today. This technique laid the foundation for our current understanding of genomes and helped to usher in a new era of genomic research.
How Early Scientists Leveraged Sanger
In the early days of DNA sequencing, the Sanger method was used to determine the order of nucleotides in a sample of DNA. This involved manually pouring thin polyacrylamide slab gels between large glass plates and a shark tooth comb was inserted to create wells for loading radioactive sequencing reactions. While these gels were very large and cumbersome, they allowed for the accurate determination of nucleotide sequences.
The Sanger chemistry used radioactive dideoxy-terminators (ddNTPs) that were initially labeled with 32P, and later with 33P and 35S radioisotopes before radioactivity was phased out and replaced entirely by the fluorescent-labeled ddNTPs that are still used today (e.g, BigDye™ Terminators).2 After polyacrylamide gel electrophoresis (PAGE) at high voltage, the glass plates were carefully pried apart, and the gel was dried down before going into a darkroom where it was brought into contact with a large sheet X-ray film to expose the film. The X-ray film was developed and placed on a light box so the researcher could manually read the DNA sequence from the sequencing ladders of the film.
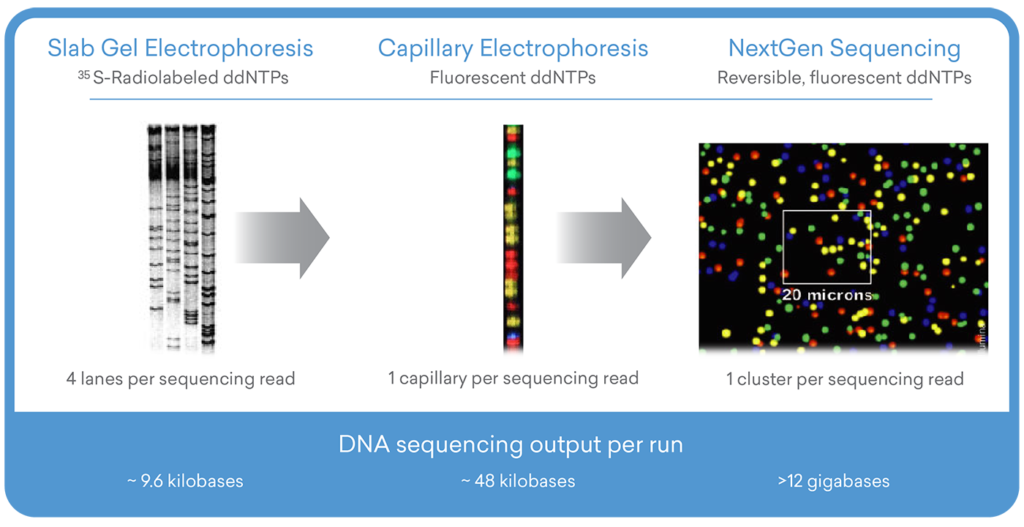
The Evolution of the Sanger Method
During the 1990s, there were several breakthrough improvements made to the Sanger sequencing method—the most notable being the introduction of fluorescent-labeled ddNTPs and the advent of capillary electrophoresis.3,4
The integration of these technologies played a critical role in the completion of the first draft human genome in 2000, a scientific achievement of such a magnitude that it is frequently compared to the first manned Apollo moon landing in 1969.5
Today’s Scientists Still Rely on Sanger’s Inventions
When Sanger developed his now-famous sequencing method in the 1970s, he likely never imagined that his chemistry would still be in use some 40 years later. But that’s exactly the case: next-generation sequencing (NGS) platforms use a modified version of Sanger’s chemistry (i.e., chain termination with reversible fluorescent ddNTPs) to achieve high throughput and accuracy.
What makes Sanger’s chemistry so special? Put simply, it is highly effective and robust. It employs high fidelity DNA polymerases to sequence long stretches of DNA with high accuracy, and it is not prone to the same kind of errors that can occur with other methods such as single molecule sequencing. This makes it ideal for applications like genome sequencing, where even a single error can have serious consequences.6
Faster, More Effective Sequencing is Possible
A reversible, fluorescent version of Sanger’s dideoxy-termination chemistry was exported from electrophoretic platforms onto NGS platforms and simply renamed “Sequencing-by-Synthesis” (SBS) chemistry. The robust chain termination chemistries originally developed by Sanger are still used today on the most advanced and successful NGS platforms in the world.
The key difference between SBS and traditional Sanger sequencing is that NGS platforms can sequence millions of DNA fragments in parallel, whereas traditional Sanger sequencing can only sequence one fragment at a time. This difference in scale is what allows NGS platforms to generate huge amounts of data in a relatively short period of time.
And that’s what we specialize in here at seqWell. Our plexWell auto-normalizing library prep technology allows the creation of balanced library pools without the need for sample or library normalization.
plexWell’s simplified 3-hour workflow multiplexes 100’s to 1000’s of samples for loading on a single sequencing run enabling enhanced overall sequencing performance. This effective, simple, and fast technology can be used for a variety of sequencing applications – from full plasmid to microbial sequencing and low-pass whole genome sequencing.
Conclusion
Three decades ago, the process of sequencing a human genome took years to complete and required a whole army of dedicated scientists. However, thanks to improvements in Sanger sequencing technologies and seqWell’s plexWell technology, it is now possible for one technician to prepare 100s to 1000s of samples for sequencing in a single day.
References
- https://www.sigmaaldrich.com/US/en/technical-documents/protocol/genomics/sequencing/sanger-sequencing
- https://bioclimate.commons.gc.cuny.edu/analyzing-dna/sanger-sequencing-of-dna/
- https://www.sciencedirect.com/topics/nursing-and-health-professions/dideoxynucleotide
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5994730
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1127709
- https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-016-0269-0

