
Introduction
Choosing the right indexing strategy can mean striking a balance between minimizing your per-sample sequencing costs and selecting the best method for your particular application and sample type.
The majority of library prep methods for Illumina sequencing incorporate dual indexing, or labeling both ends of the library molecules with barcodes to enable pooling of many samples into a single flow cell for more economical sequencing.
In this blog, we’ll discuss the differences between unique dual indexes (UDIs) and combinatorial dual indexes (CDIs), and which applications or sample types may benefit most from the use of UDIs.
UDI versus CDI: What’s the Difference?
In dual indexed library preparation, two different index barcodes are added to the library via barcoded adapters or PCR primers: the “i7” index is added on one end and is typically read first, and the “i5” index is added on the other end and read second.
In both CDI and UDI, each sample within a pool is identified by the combination of i7 and i5 indexes. In CDI, each i7 and/or i5 may be used in a combinatorial fashion, in other words the combinations are unique but each i7 and i5 is used multiple times within the set. In contrast, UDIs have completely unique i7 and i5 pairings and each is only used once within the set.1,2 For example, a set of eight i7 indexes and eight i5 indexes can be combined to generate a set of 64 total CDI combinations, but can only be combined to generate a set of 8 UDI combinations.
Using UDIs to Mitigate Index Hopping
The use of CDIs can be highly desirable for many applications and sample types because it more easily enables high scale multiplexing of up to thousands of samples per sequencing run. However, one of the main uses for UDIs is to mitigate the effects of “index hopping” that occurs in Illumina’s patterned flow cell chemistry used on the NovaSeq, NextSeq 1000/2000, HiSeqX/3000/4000, and iSeq sequencing instruments.3,4 In these instruments, this hopping phenomenon likely occurs due to excess index primers or adapters in library pools interacting with the unique patterned flow cell chemistry, which can lead to extension of library fragments with an oligo containing another sample’s i7 or i5 index.
This hopping generally occurs at a rate of <1% of total reads for most library methods. However, patterned flow cell chemistry is not the only place where index hopping can happen; one can expect to see low levels of index hopping in any multiplexed or pooled PCR where multiple indexed samples are amplified in the same well or tube.
If you are using indexes combinatorically, these “hopped” index reads cannot be easily identified and could be attributed to the wrong source sample during demultiplexing. Using UDIs provides a unique two index signature for each sample, and both the i7 and i5 index must match an expected combination. Any reads with an unexpected i7 + i5 combo can be removed during standard demultiplexing. It should be noted that for this reason, the use of UDIs will lead to a higher rate of discarded data compared to CDI.
While most applications may be tolerant of low levels of read misassignment between samples, certain applications may be more sensitive and can benefit from using UDIs. These typically are the “needle in a haystack” type of projects where a high sensitivity or limit of detection is required. Some examples include low allele fraction tumor sequencing, detecting rare transcripts or fusion events in RNA-seq, or identifying the presence or absence of low abundance bacterial strains in microbiome samples. Additionally, some labs may feel more comfortable using UDIs versus CDIs across the board, particularly for core or service labs that process many different sample and project types or have patterned flow cell sequencers.
Normalizing UDI Library Construction
Most available UDI methods incorporate indexing into the library PCR step, which can be labor and cost intensive.
One approach to alleviate these challenges is by using an auto-normalizing UDI library construction method that permits pooling of samples immediately after sequential transposase mediated tagging steps.5
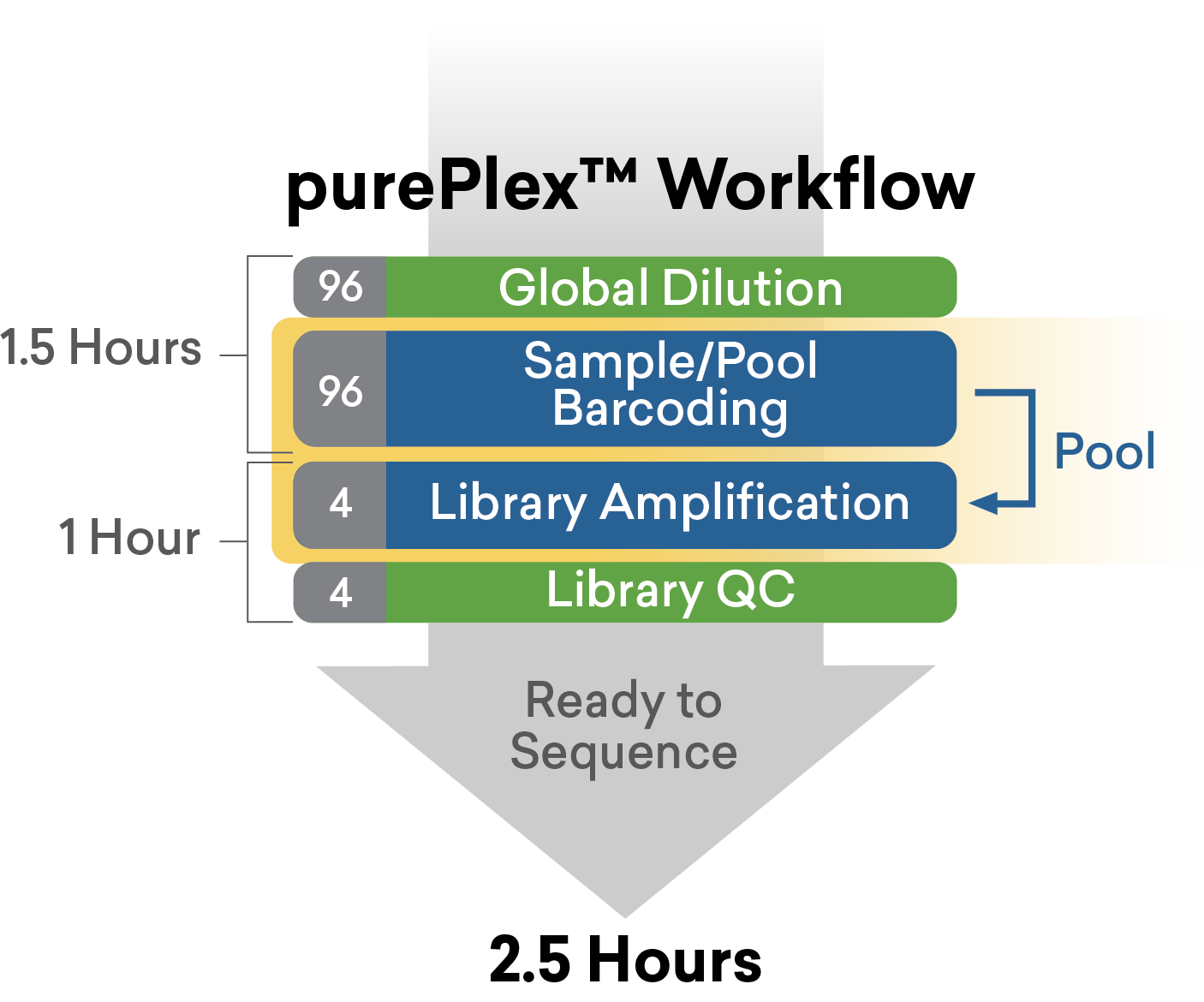
In our purePlexTM DNA Library Preparation workflow, auto-normalization is achieved through sequential transposition events of full-length indexed adapters in the presence of a novel normalization reagent. This approach allows incorporation of unique dual indexes and permits pooling of samples immediately following the tagging steps such that purification and amplification of fragments occur after pooling, reducing the QC and labor costs compared to traditional UDI workflows.
A distinct advantage of the purePlex DNA library preparation workflow is the elimination of individual sample and library normalization. 5
Summary
CDIs and UDIs each have applications for which they may be better suited. Applications involving massive levels of multiplexing benefit from the use of CDIs due to the ability to easily reach index combinations in excess of 384. In contrast, UDIs are often the optimal strategy for applications of the highest sensitivity that do not require routine high levels of multiplexing. Still, many applications sit between these two extremes and could be well suited to either CDI or UDI library preparation strategies.
References
- https://www.illumina.com/techniques/sequencing/ngs-library-prep/multiplexing/unique-dual-indexes.html
- https://support.illumina.com/bulletins/2018/08/understanding-unique-dual-indexes–udi–and-associated-library-p.html
- Minimize index hopping in multiplexed runs.
- Costello M et al. (2018) Characterization and remediation of sample index swaps by non-redundant dual indexing on massively parallel sequencing platforms. BMC Genomics 19:3
- https://seqwell.com/normalizing-udi-library-construction-poster/
