Transposase-based NGS Library Preparation
Harnessing the Power of Transposases
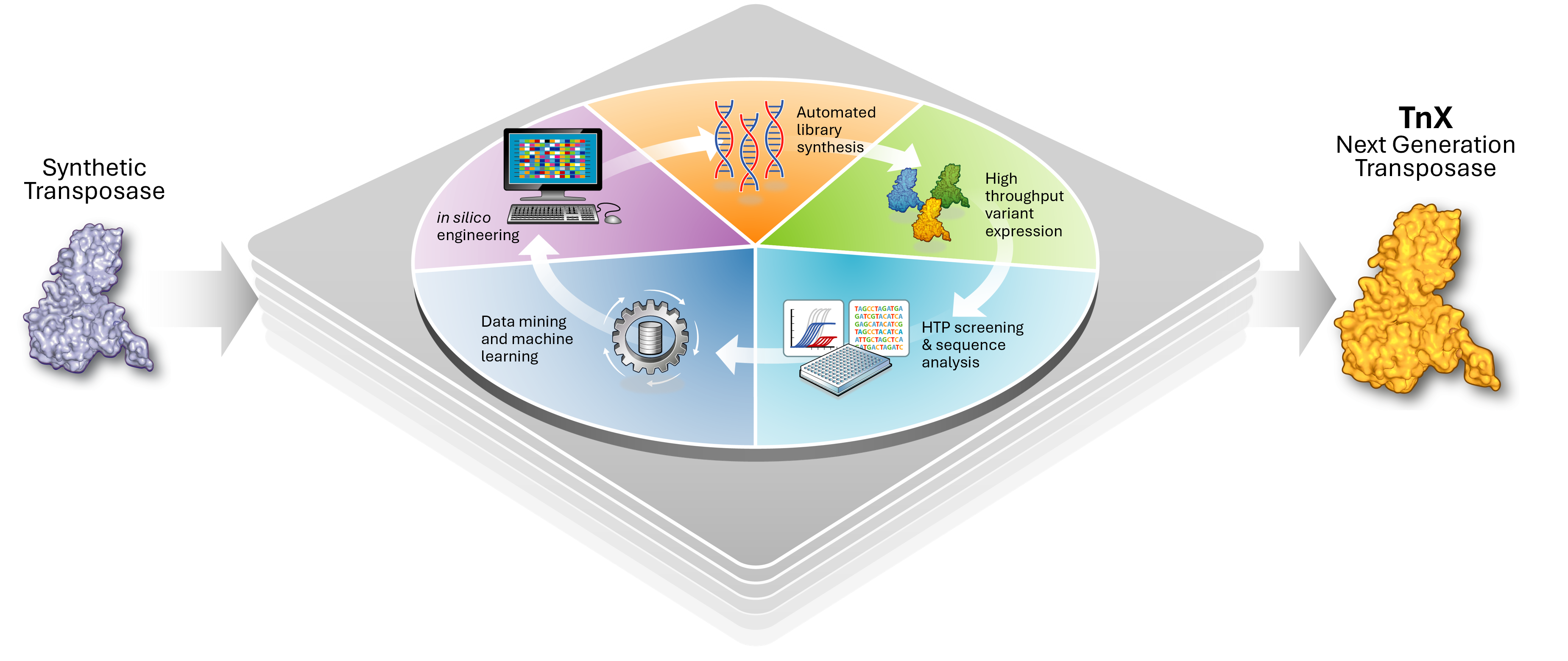
seqWell innovation began in 2014 by harnessing the power of tagmentation – the simultaneous fragmenting and tagging (ligating NGS adapter sequences) of DNA – to replace traditional library preparation methods that use complex and lengthy protocols and to increase the power of multiplexing. Our latest innovations include TnX, an engineered next-generation transposase and high-performance alternative to Tn5, and seqWell directional tagmentation.
seqWell uses a variety of NGS library preparation workflows — from one-step tagmentation & amplification, to single and dual tagmentation, as well as directional tagmentation — to address a variety of needs.
- NGS system compatibility: Illumina, PacBio, Element Biosciences, Complete Genomics & Ultima Genomics
- Sequencing technology: Short-read and long-read sequencing
- Throughput: From 10’s to 1000’s of samples
- Sample types: Plasmids, amplicons, RCA, colony PCR & genomic DNA
- Kits to reagents: Ready-to-use kits and custom-loaded transposases
TnX… don’t be so biased
Engineered for Performance
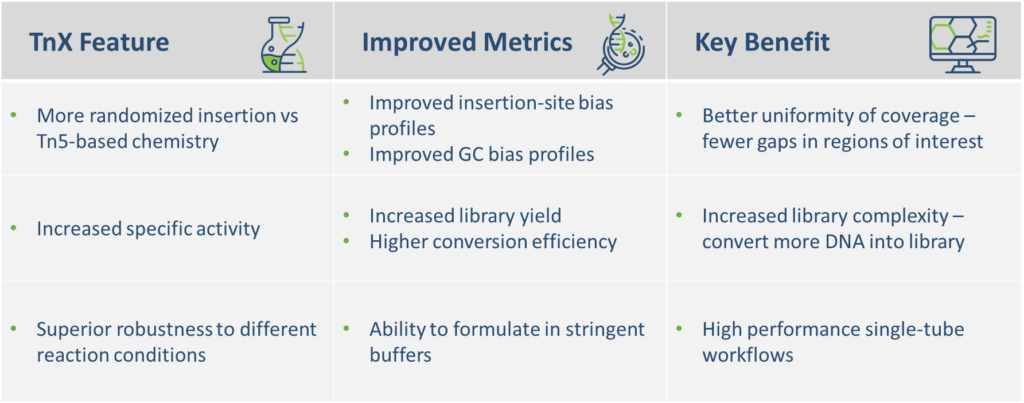
TnX, seqWell’s next generation transposase and alternative to Tn5, was developed using fit-for-purpose engineering that targeted improvements in 3 key enzyme attributes: activity, insertion bias and robustness. These improved attributes translate into enhanced sequencing performance by improving NGS data quality and uniformity.
Better transposase, better libraries, better NGS
Reduced insertion site bias
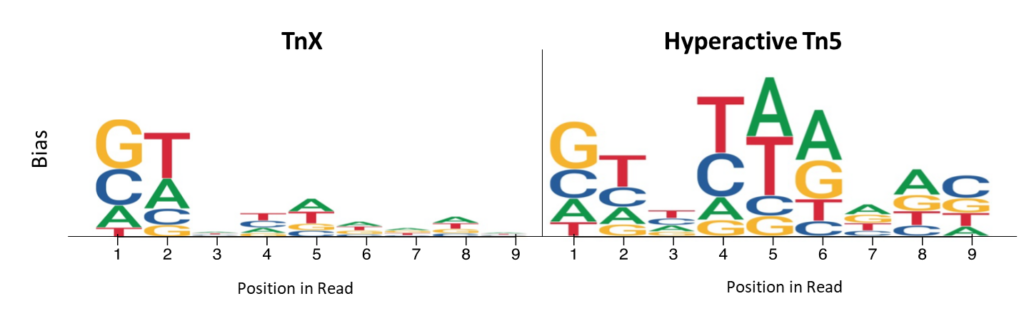
Read start site insertion bias was measured by examining the frequency of bases in the first 9 bases of each read. Positions with higher per-base nucleotide bias are represented by heights for hyperactive Tn5 and TnX, and illustrate the reduced bias of TnX.

Products Powered by TnX
-
Tagify Custom-loaded Transposase
TnX loaded with universal adapters, i5/i7 adapters, or customer-specified oligos – used for sensitive targeted gene editing QC analysis assays, as well as other custom assays.
-
ExpressPlex 2.0
Ready-to-use, one-step library preparation kit for plasmid, amplicon and small genome sequencing
-
MosaiX (early access)
90-minute library prep using TnX-based directional tagmentation for production of high complexity libraries.
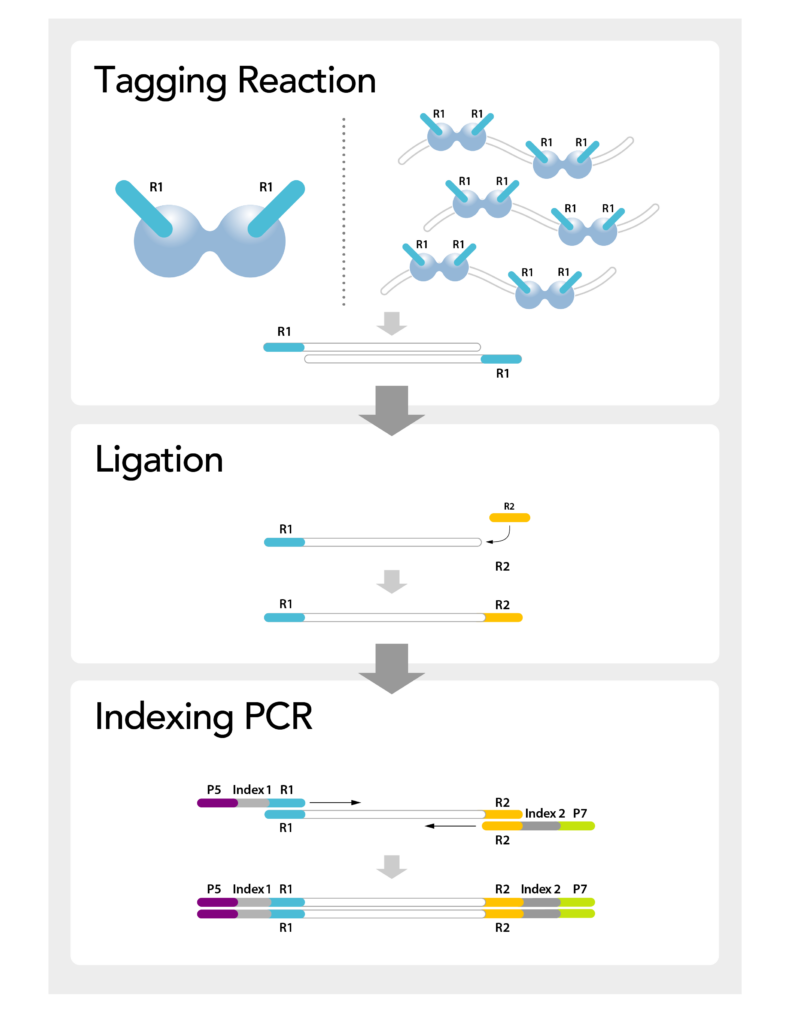
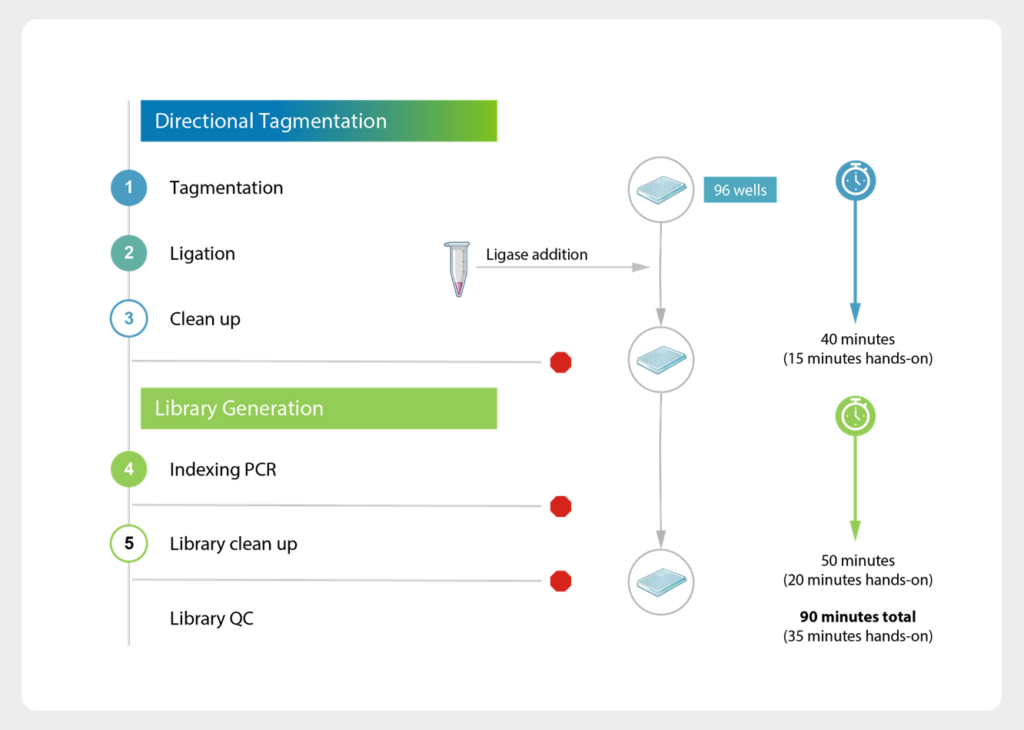
seqWell directional tagmentation… complexity made simple
The Ease of Tagmentation, with the Performance of Ligation
Combine the workflow simplicity of tagmentation, the reduced insertion bias of TnX, & the performance benefits of ligation and you’ve got seqWell’s directional tagmentation used in our ready-to-use MosaiX kit. This 90-minute workflow requires less than 35 minutes of hands-on time and produces complex libraries required for population genomics.
Blogs of Interest

PhiRx™: A Color-Balanced Spike-In Control for Improving Demultiplexing on XLEAP-SBS Illumina Sequencing Instruments

Short-Read Sequencing vs. Long-Read Sequencing: Which Technology is Right for Your Research?

Novel tagmentation-based workflow that improves throughput and scalability of long read assays
seqWell™ Library Preparation Kits and Reagents
Our library prep and multiplexing kits are available to support all your short and long-read sequencing applications. Contact us to find out which is right for you!