SARS-CoV-2 surveillance sequencing with plexWell technology

RECORDED WEBINAR
Caught in the Act: Tracking the Emergence and Divergence of SARS-CoV-2 through Statewide Testing and Sequencing
Simple workflow
![]()
ARTIC-compatible with no upstream purification
High throughput
![]()
Multiplex thousands of samples
Uniform Coverage
![]()
Down to 160 viral copies
We need to understand the SARS-CoV-2 virus now, and NGS provides a scalable approach to quickly analyze thousands of specimens.
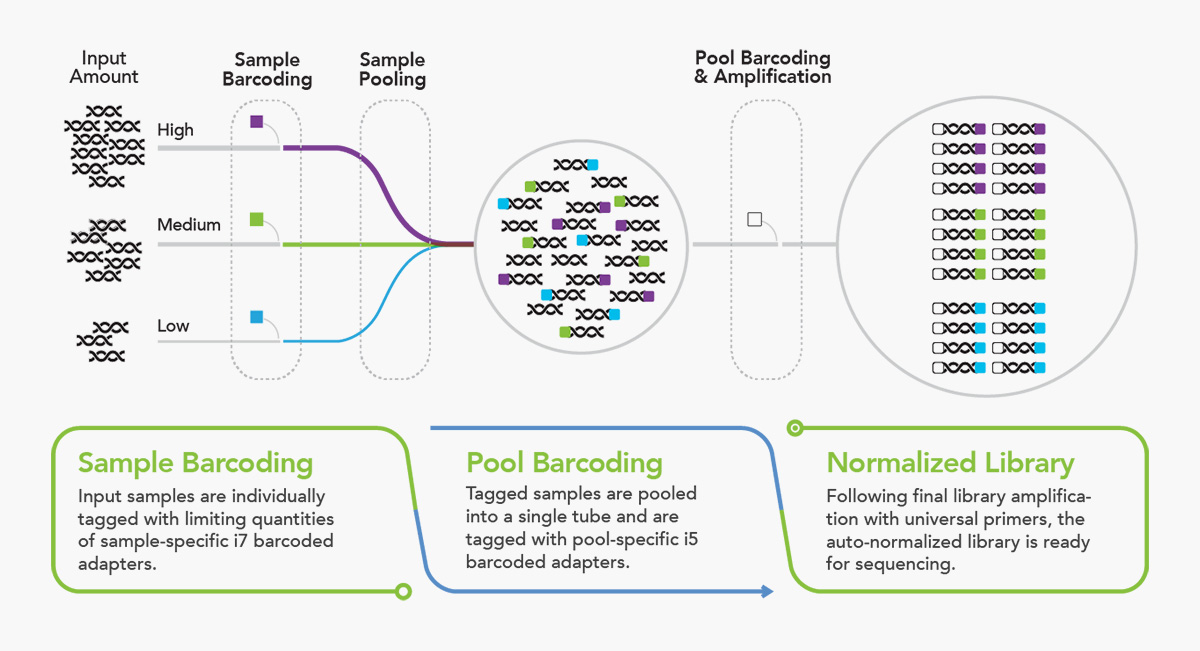
Current technology, though, is limited by the number of samples sequenced simultaneously (multiplexing), sensitivity, and turnaround time. plexWell™ technology combines a simple and efficient workflow with high sensitivity and unparalleled scalability that is ideal for high-throughput SARS-CoV-2 sequencing.
Efficient and Highly Multiplexed Whole Viral
Genome Sequencing of SARS-CoV-2 Samples
Our new plexWell sample preparation method in combination with a widely-used ARTIC RT-PCR protocol greatly simplifies and increases the throughput of multiplex whole genome sequencing of SARS-CoV-2 RNA. It provides high quality results while reducing the time and cost of sequencing efforts. The method provides numerous advantages, including:
• robust performance
• simple workflow
• high multiplexing capacity
This cost-effective solution can provide up to 40% savings compared to other methods and reduce processing time from sample to result. The plexWell workflow is automation-friendly and compatible with a variety of platforms. plexWell enables sequencing of up to 2,304 samples in a single sequencing run and provides a simple workflow capable of meeting the high demand for COVID-19 testing.
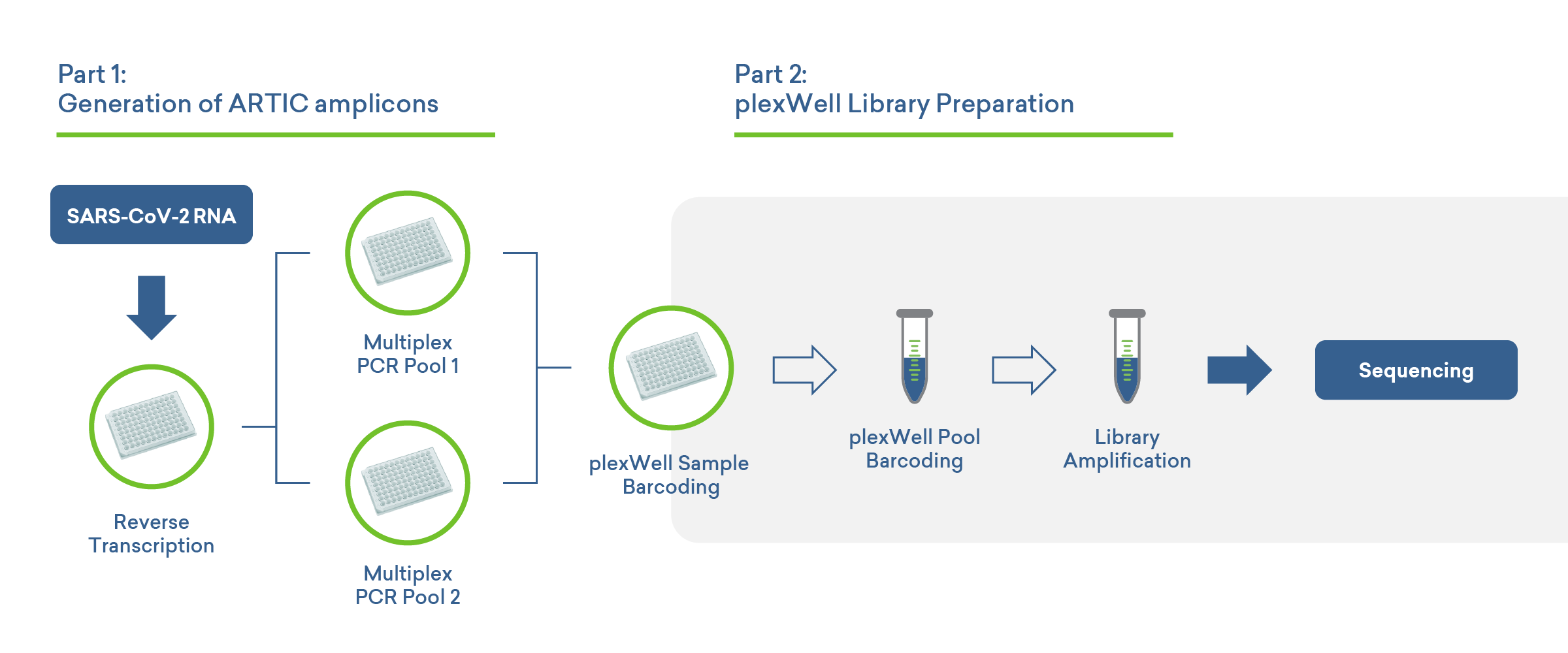
SARS-CoV-2 sequencing using plexWell
This protocol describes how to generate NGS libraries for whole genome sequencing of SARS-CoV-2 isolates, combining methods for RT-PCR from the ARTIC Network for generating tiled amplicons from coronavirus RNA [1] along with downstream library prep with standard plexWell multiplexed NGS library prep reagents.
Streamlined workflow for SARS-CoV-2 whole-genome sequencing
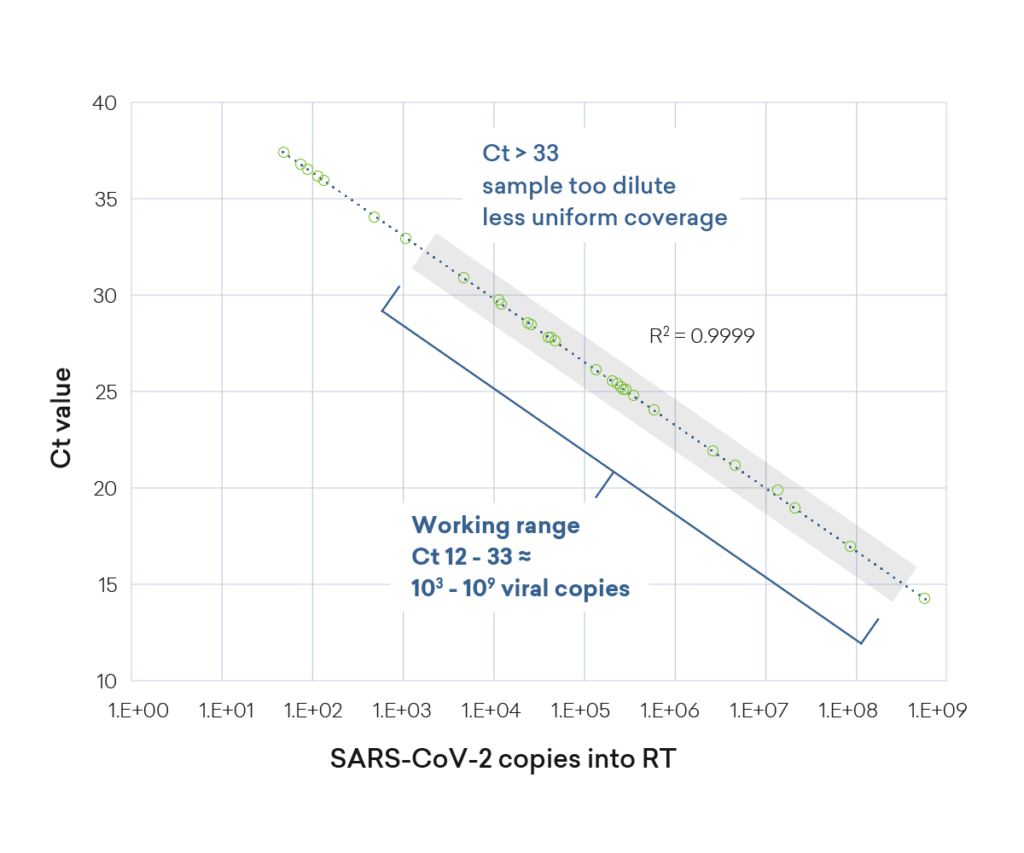
Viral load range in clinical SARS-CoV-2 clinical specimens
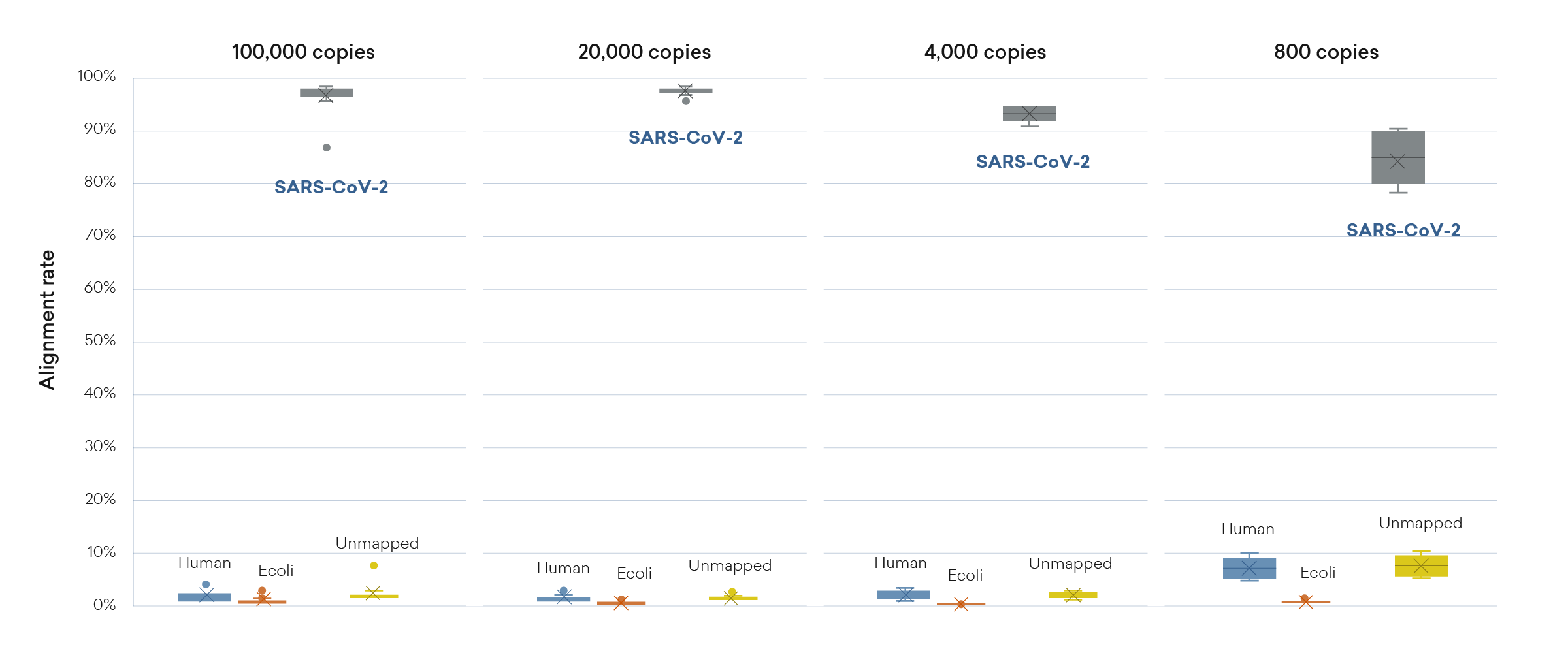
SARS-CoV-2 viral load varies widely in nasal swab samples that are positive for COVID-19, and plexWell assays have the sensitivity to work with all but the most dilute virus-positive samples. Total nucleic acid isolated from saliva was used as the input, with the number of input viral genome copies similar to what is found in a typical nasal swab sample (~4.2 x 104 viral copies). Following RT and multiplex PCR, libraries are prepared using the plexWell kit. The libraries were then sequenced to produce full-length SARS-CoV-2 sequences.
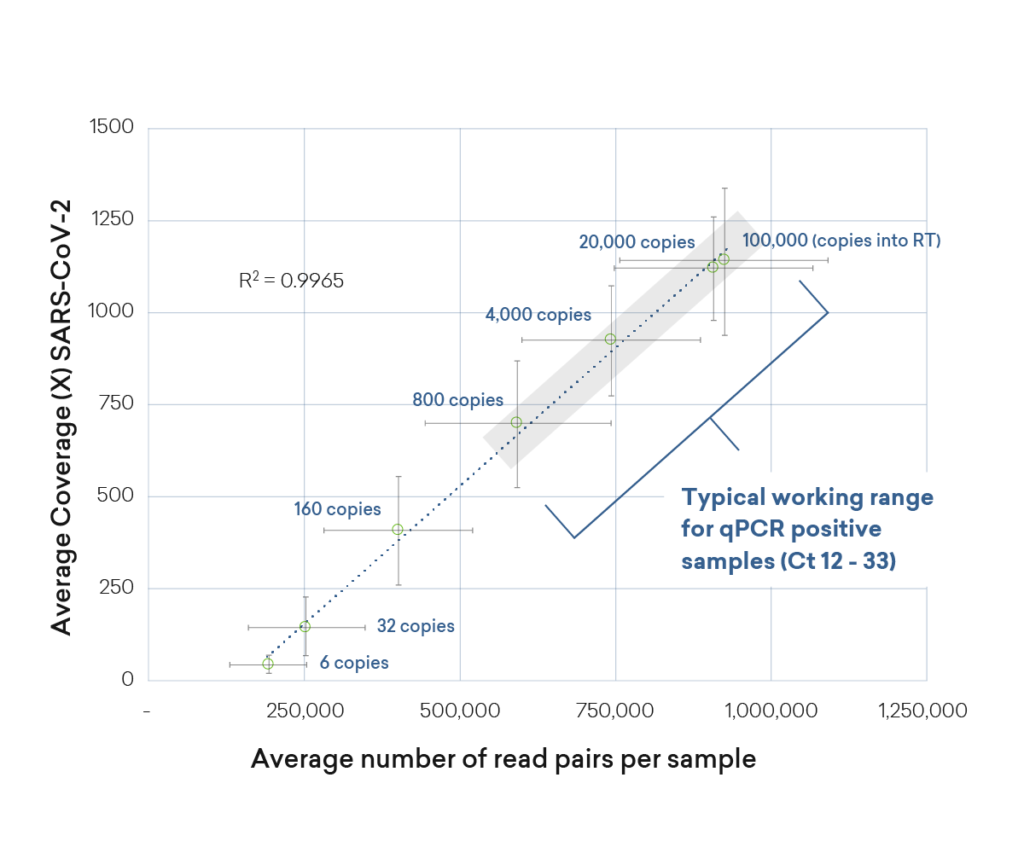
Uniform and high SARS-CoV-2 coverage
The plexWell library prep kit delivers uniform and high sequence coverage for even low-input SARS-CoV-2-positive samples. Even at low copy numbers (160 copies), plexWell delivered 400X coverage with strong uniformity.
High SARS-CoV-2 accuracy
The plexWell library prep kit produces libraries with high mapping rates even for low-copy number SARS-CoV-2 samples.