Improved library workflows for quick turnaround of plasmid, amplicon, or synthetic construct sequencing
for quick turnaround of plasmid, amplicon, or synthetic construct sequencing
Plasmids and PCR products are the bread and butter of molecular biology labs all over the world.
Broad-based access to cost-effective, next-generation sequencing (NGS) and advances in synthetic biology have revolutionized the scale to which plasmids can be constructed, screened, and manipulated for wide-ranging applications, including biomarker discovery, antibody engineering, genome editing, and gene therapy. However, NGS library preparation throughput, performance, and cost, remain bottlenecks in high-throughput plasmid sequencing.
Amplicon sequencing is a targeted NGS approach that enables researchers to analyze genetic variants in specific regions of a genome. This technique is particularly useful for detection of hot-spot mutations, copy number variations, gene fusions, insertions, deletions and single nucleotide polymorphisms (SNPs). Because of the sensibility and the versatility of amplicon sequencing, efficient library preparation is a must.
Our library preparation kits offer robust workflow solutions for amplicon sequencing by focusing on scalability (high-throughput multiplexing), speed (as fast as 90 minutes with some kits), and cost-effectiveness. Our workflows are easy to implement and automation-friendly. We provide improved yield across a broad range of sample types in various disciplines, including precision medicine, gene therapy, and gene editing.
Simple workflow
![]()
Significant reduction in normalization and library QC time
High throughput
![]()
Pooled multiplexed library prep without sacrificing quality
COST EFFECTIVE
![]()
30-50% reduction in labor and consumable costs

RECORDED WEBINAR
Moderna presentation – Antibody Sequencing: New Operating Model
In this presentation, Nasthas Lacerda Almeida from Moderna, highlights how her organization has leveraged ExpressPlex™ 2.0 for a clonal antibody sequencing approach, optimized for speed and scalability using de novo assembly of NGS short reads.
Our transposase-based technology removes critical bottlenecks in NGS workflows by enabling scientists to prepare samples into libraries and get them onto sequencers quickly and with higher performance.
Current technology is limited by the number of samples sequenced simultaneously (multiplexing), sensitivity, and turnaround time. seqWell’s technology combines a simple and efficient workflow with high sensitivity and unparalleled scalability that is ideal for applications such as high-throughput plasmid and amplicon sequencing, and for use in gene therapy and sythetic biology.
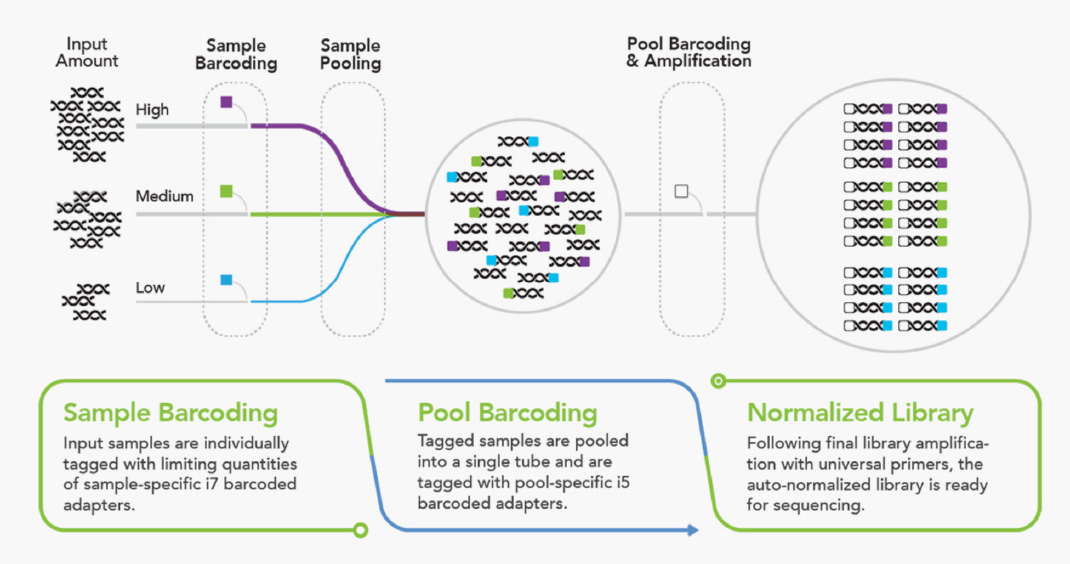
Download our Application Note to discover how our plexWell Library Preparation Kits outperform competitor transposase-based workflows in terms of ease-of-use, flexibility, and overall data quality for high-quality, high-throughput plasmid sequencing on benchtop sequencers.
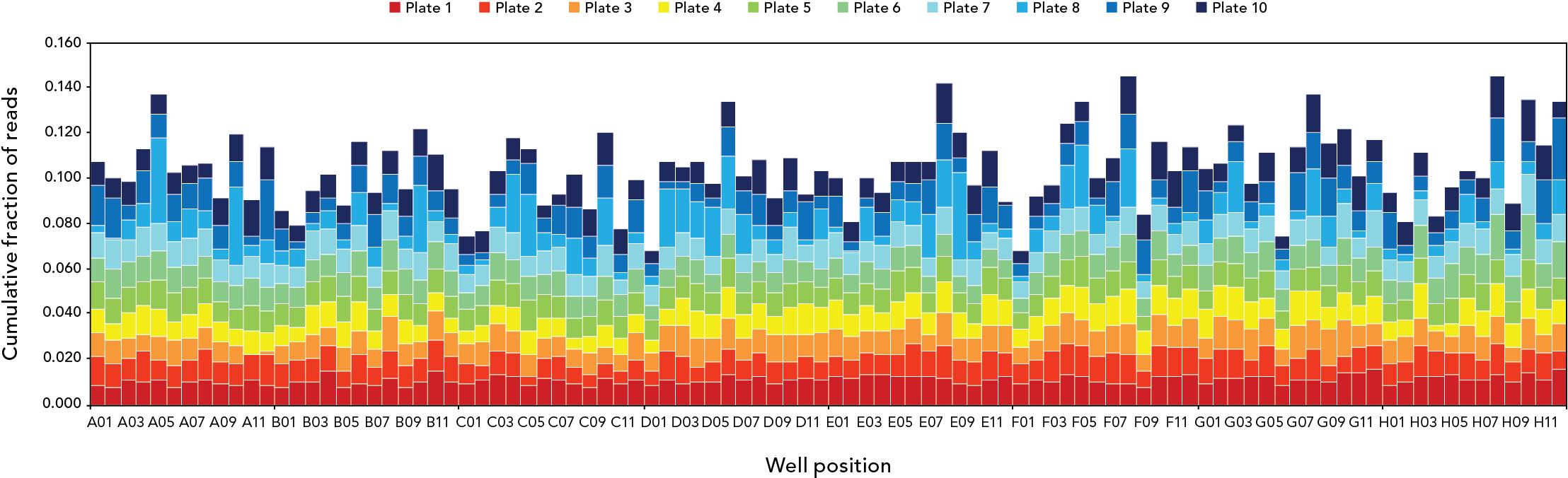
Read distributions by plate and well position for 960 plasmids sequencing in a single Illumina® MiSeq™ (2 x 250 bp) run. Samples were processed in 10 plates of 96 plasmids each with the plexWell 384 Library Preparation Kit. Assemblies were obtained for 955 of the 960 plasmids (99.5%), and were free of ambiguities for 99.2% of the samples.
Our cost-effective NGS workflow solutions can provide 30 – 50% savings compared to other methods and reduce processing time from sample to result.
A crucial technology that we use to build almost all of our products is an enzyme called Tn5 transposase, which is an incredibly versatile tool for scaling and simplifying NGS workflows. We have found Tn5 to be an incredibly powerful platform for creating library chemistry with several unique advantages, such as auto-normalization and multiplexing with purpose-built reagents. These reagents form the core capabilities that our products are based on.
The following products support specific applications like plasmid and amplicon sequencing, synthetic construct sequencing, microbial genomics, and metagomics/microbiome screening, in both research and commercial settings.
ExpressPlex™ 2.0 Library Prep Kit
This kit was designed for applications that focus on speed and volume versus high-depth reads. For example, if your lab is transitioning away from Sanger because you need to process at larger volumes, ExpressPlex 2.0 can dramatically shorten time to data. ExpressPlex 2.0 is one-step tagmentation-based library preparation method that is automation-friendly and extremely scalable.
Learn More About ExpressPlex 2.0
plexWell™ 96 or 384 Library Prep Kit
The plexWell technology allows multiplexed libraries to be prepared as a single pool with significantly improved cost, time, and performance. It normalizes across a wide range of input DNA amounts through Tn5 transposase-based chemistry. The plexWell workflow is automation-friendly and compatible with a variety of platforms.
The method provides numerous advantages, including:
• Robust performance
• Simple workflow
• High multiplexing capacity
Tagify™
Tagify is a versatile Tn5 transposase-based toolkit for custom NGS library preparation. We have a wide range of tagging reagents available.
Applications include:
- Single-cell ATAC-Seq
- CRISPR edit verification (UDiTaS method)
- Cell/gene therapy
- Single-cell RNA-seq
For more information on how we have leveraged the versatility of the Tn5 enzyme to help scale NGS workflows, check out our blog, “Enabling Sequencing Applications with Improved Transposase-Based Solutions.”
Featured Resources for Plasmid and Amplicon Sequencing
APPLICATION: CRISPR/Cas9 QC
This 2022 publication, “Barcoded bulk QTL mapping reveals highly polygenic and epistatic architecture of complex traits in yeast”, explores barcoding bulk quantitative trait locus mapping. The paper discusses strategies to increase the scale of the study by use of combinatorial index strategies, minimization of pipetting and purification steps, and use of seqWell’s transposase-based reagents that incorporate index adapters directly into the samples.
APPLICATION: Amplicon Sequencing (Viral)
In this publication by Samer, et al., “Blockade of TGF-β signaling reactivates HIV-1/SIV reservoirs and immune responses in vivo”, our plexWell 384 kit was used to prepare library from gag gene amplicons.
The plexWell 384 kit contains four assay-ready, 96-well PCR plates that are fully-skirted and low-profile and offers barcode combinations to multiplex up to 2,304 samples in a single lane.
APPLICATION: High-throughput Screening
New England Biolabs multiplexed sequencing libraries using our plexWell 384 Library Preparation kit in the study, “Capillary Electrophoresis-Based Functional Genomics Screening to Discover Novel Archaeal DNA Modifying Enzymes”.
The authors reported that plexWell 384 provided “a rapid, simple, high-throughput method to discover novel archaeal nucleic acid modifying enzymes by utilizing a fosmid genomic library, next-generation sequencing, and capillary electrophoresis.”
SCIENTIFIC POSTER: Multiplexed phasing of clinically relevant long human PCR amplicons with short reads
We have developed a novel library preparation technology that links short reads across long DNA fragments and multiplexes many samples for fast and affordable parallel phasing of dozens of gene-length amplicons.
SCIENTIFIC POSTER: Multiplexed Transposase-Based NGS Tools for QC Assays in Genome Editing Applications
We describe the application of a customized Tn5 transposase (Tagify™ i5 UMI TR) to characterize and quantify transgene induced editing in the human genome, and the application of the UDiTaS technology to assays of genome editing of the clinically relevant human gene CEP290.
SCIENTIFIC POSTER: Comparison of a Transformational One-Step Library Prep Method for High-Throughput, Multiplexed Sequencing
This poster showcases seqWell’s transformational library prep technology that enables barcoding and amplification of 8 – 384 dual-indexed libraries in 90 minutes in a “one-pot” reaction, while simultaneously auto-normalizing library read count and insert size.
