
Introduction
A vital factor in NGS is control over sample mass input, which is the amount of sequenceable molecules that are produced during any given reaction.
Auto-normalization at the beginning of a library prep workflow allows researchers to save substantial time and resources. To this point, variable mass inputs that yield similar sequencing read outputs are beneficial as they save researchers on the time required for QC, the costs of sequencing, and the amount of samples that need to be loaded.
More total mass input is often favorable with traditional ligation-based library prep methods because it increases the complexity and unique molecules of a library, which increases the assay’s detection sensitivity.1 For this reason, if there is abundant mass, a researcher might load more onto their sequencer in an effort to obtain a more complex data output for their study.
However, does increasing mass in an auto-normalizing, Tn5 transposase-based library prep workflow function the same as with traditional methods? Is more sample mass input always better?
A Primer on Variable Sample Mass
Let’s take this one question at a time. To answer the first, we must outline the effects of variable sample mass.
In transposase-based library prep, the number of insertions of the limiting P7 barcode remains the same. However, the distance between those insertions is directly proportional to the amount of mass one puts in. This is a result of the fact that the reagent is limiting, and the entirety of it will be used in the initial reaction.
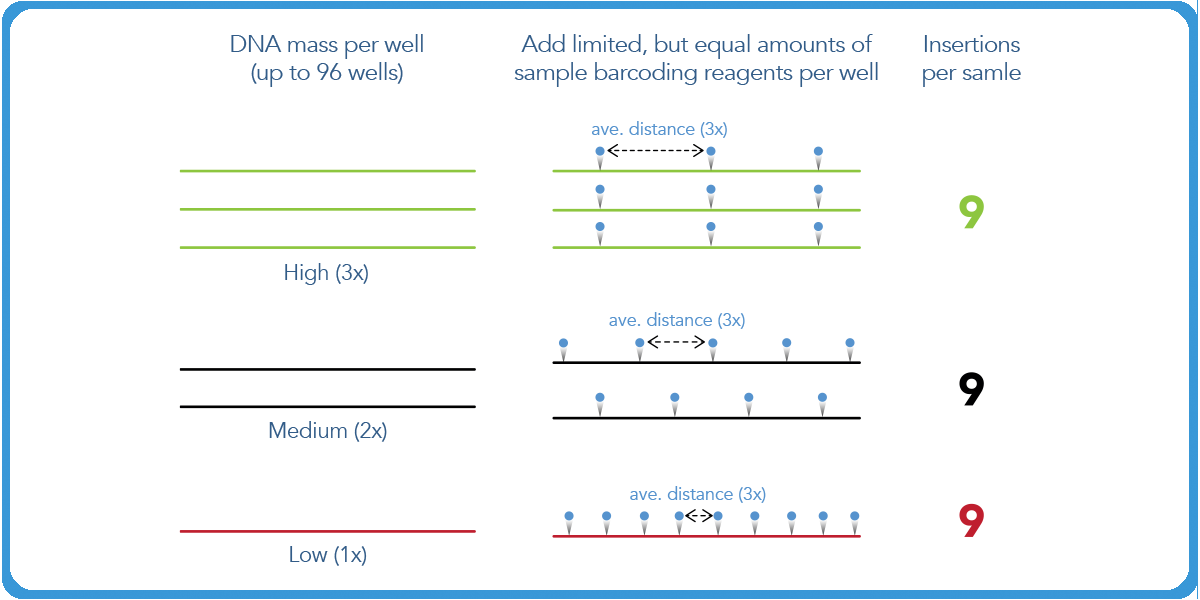
The second P5 index is then added in excess. The insertions will occur at the same optimal distance from each of the P7 indices. Then a PCR reaction is conducted with P5 and P7 primers. As a result, the distance between P5 and P7 is the length of the final library.
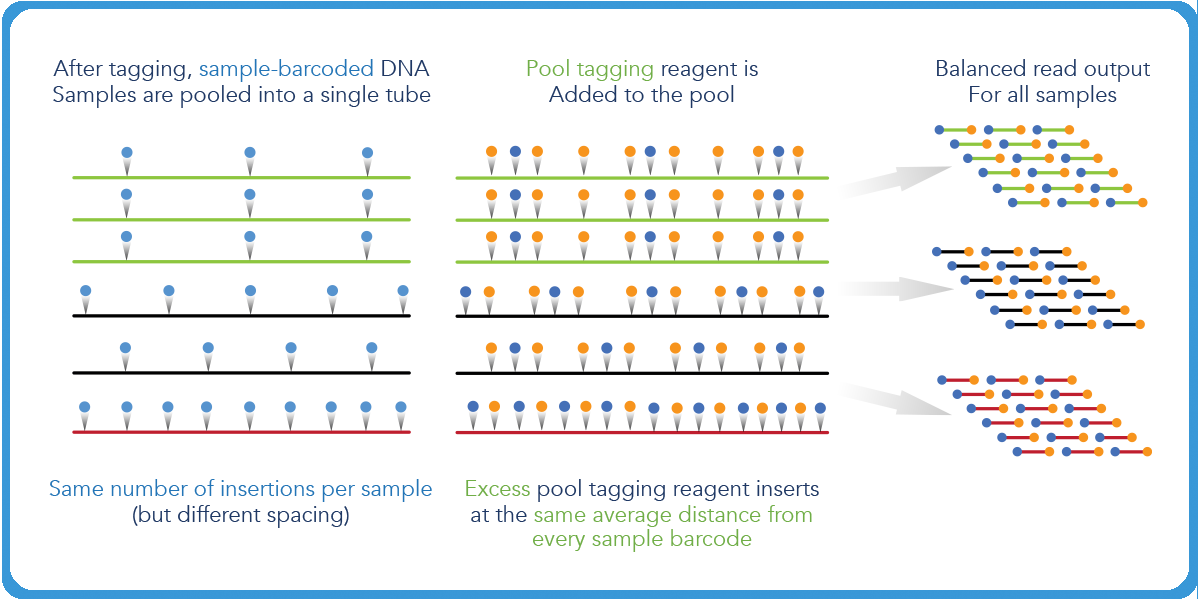
When more mass is inputted in this reaction, the distance will shift to a larger degree, which will have an impact on the final sequenceable molecules of the whole pool.
So Is More Always Better?
Researchers often think that more sample input mass might solve their library prep woes. However, once we understand the impact of mass on different types of workflows, we realize that’s not always true.
With normalizing Tn5 transposase-based reagents, the effect of more total mass is proportionally related to library length. This is a crucial consideration as longer fragment lengths can be a major driver in increased error rates.2 In contrast, shorter library fragments cluster more efficiently than longer libraries, so a shift to a longer library length would lead to less data output.3
If a researcher was trying to increase the amount of reads per sample, which is often the reason for adding more mass, this could have the entirely opposite effect than what they intended.
The Power of Transposase-Based Library Prep
Now, although a change in total mass input across samples could have debilitating effects when implemented incorrectly, there are still a plethora of advantages to using transposase-based library prep over traditional ligation-based methods.
Transposase-based library prep gives researchers control over their transposition reactions.4 Auto-normalization, which researchers can leverage with the plexWell™ 96 and 384 library prep kits, ensures uniform insert size, even in cases of variable sample inputs so long as the average and total input of all samples stays the same.
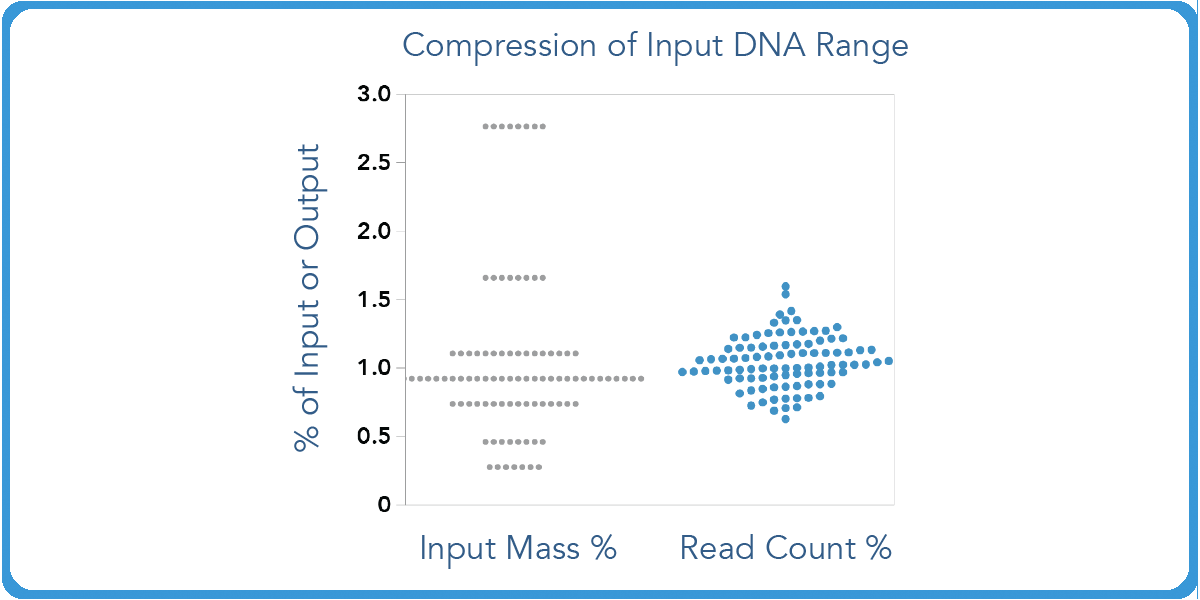
plexWell also compresses read count distribution over a 10-fold input range. So users can rest assured that they will maintain a consistency no matter the percentage of input or output mass.
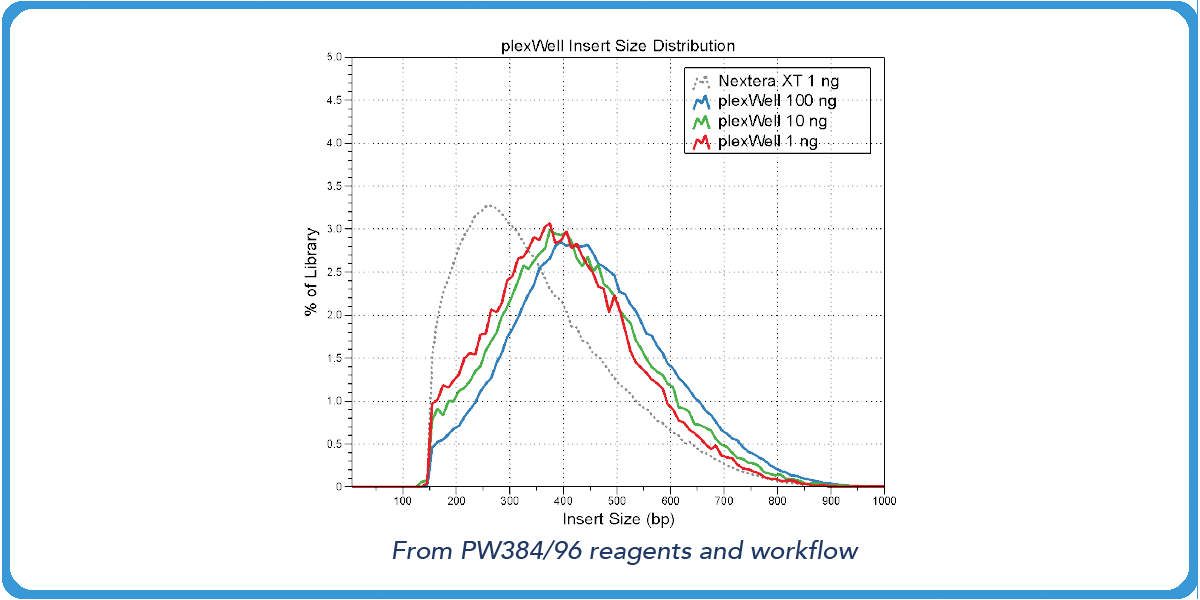
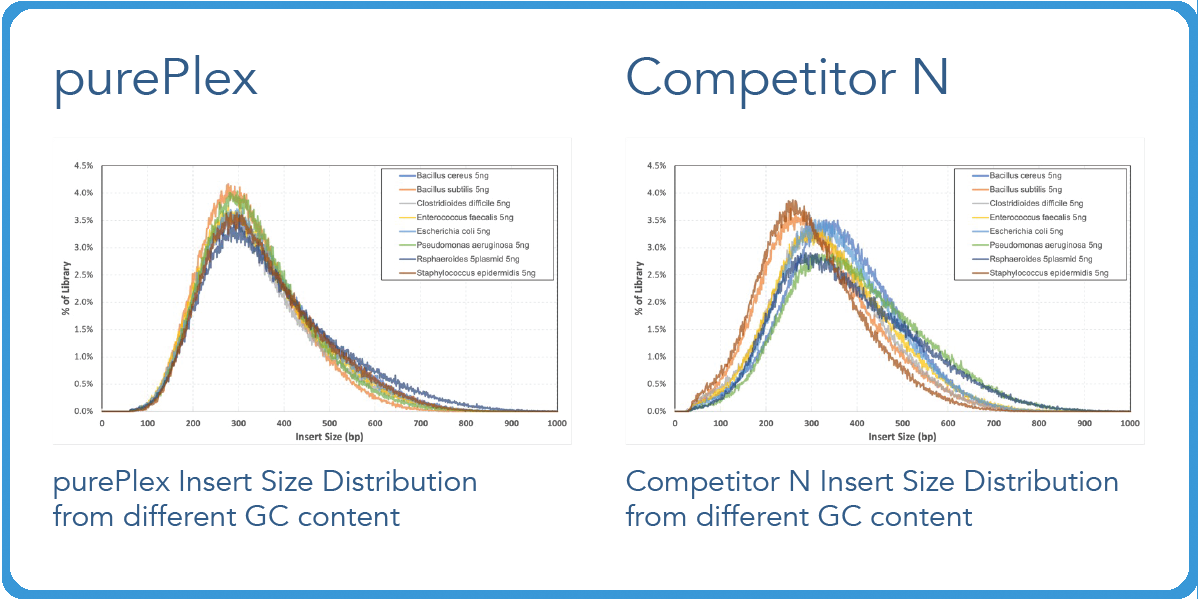
Transposase-based library preps from competitors have varied fragment distributions from GC content even after normalizing the sample input.5 However, with our purePlex™ DNA Library Prep Kit, insert size within a pool of samples is consistent regardless of the input (panel A) or GC content (panel B).
Conclusion
Variable mass inputs that yield similar sequencing read outputs are beneficial as they allow for a similar quality and coverage across libraries. This remains true even with variations in the amount of extracted samples.
However, regarding transposase-based library prep, more input mass is not always better. Innovations with auto-normalization, such as the plexWell and purePlex library prep kits, ensure consistency across a variety of input ranges so researchers won’t be stalled by the shifts in fragment length that result from inputting more mass into their reactions.
References
- Journal of Molecular Diagnostics | Impact of reducing DNA input on NGS
- Science Direct | Long fragments achieve lower base quality
- Illumina | How short inserts affect sequencing performance
- seqWell | Tn5 Transposase: A Breakthrough Enzyme for DNA Library Preparation
- Frontiers in Microbiology | Varied fragment distributions
