Introduction
This article is part of an ongoing series that highlights seqWell’s multimedia content offerings. The following is a transcript of our webinar: Driving High-Throughput NGS Library Prep With Automated Liquid Handling.1
Curtis Knox, Director of Product Management at seqWell, was joined by Paul Lomax, Head of Genomics at SPT Labtech. They opened the presentation with the current state of synthetic biology and needs for that market. Curtis and Paul also explored the challenges facing researchers in synthetic biology discovery, offering unique perspectives based on their individual areas of expertise, before highlighting the collaboration between seqWell and SPT Labtech. Then the two discussed the key benefits of the ExpressPlex Library Prep Kit and firefly automated liquid handler, and how, when used in tandem, enable better and higher throughput library preparation.
Current State of Synthetic Biology
Paul Lomax: Welcome and thanks for joining the discussion. This all started at the beginning of 2023 when we first talked to Curtis and seqWell. Back then, Curtis had some clear ideas about what was needed for synthetic biologists which he’ll talk about now.
Curtis Knox: Yes, a lot of my focus is on synthetic biology and what I wanted to try to tackle with SPT was around how synthetic biologists and researchers do their discovery today. What that comes down to is a tremendous amount of what I call “rinse-and-repeat”. In other words, you need to make new constructs, sequence those, analyze them, and then turn around again once you’ve actually looked at what you’ve created and say, “Okay, well now I need to do this or do that, or I didn’t have what I really needed.” It tends to be a very repetitive process and the quicker and easier that you can work through this type of system where you’re trying to continuously construct and analyze, the quicker you can get to your discovery.
So our idea here is how can we go about and make this process simpler for those that are doing it? And one of the things that we really thought about was, “Well, okay, so we can work with SPT in order to automate some of these systems.” And here’s where Paul will come in to talk a little bit about what systems might be useful and how we went about thinking what might make the most sense.
An Array of Automated Liquid Handlers
Paul: Thanks, Curtis. So yeah, the really interesting thing about the ExpressPlex protocol is it’s beautifully simple and it’s really great for automation.

As you can see from the image above, SPT Labtech has a number of products that could all actually be used to run the protocol to some extent. And it was a case of trying to decide which was the best platform to do that.
Our products range from simple semi-automated 96 and 384-well pipetting systems through to non-contact dispensing systems. We’ve also got ultra-low volume positive displacement pipetting systems. And then we’ve got more walkaway platforms which have a higher level of automation. And within these, there are a few core technologies that we employ.
We’ve got all these weapons in our arsenal, but ultimately we settled on using the firefly, which is our most recent addition to our liquid handling portfolio and kind of an all-in-one platform. So this actually combines a 96 and 384 pipetting head, and it also has non-contact dispensing along with incubation and shaking, and a really simple intuitive user interface.
The main reason we chose this platform was that it was great for the whole plate transfers that we needed for the high throughput in this workflow. It can also work with a single column of tips, which lent itself to the pooling part of the protocol that I’ll show a little later. We also acknowledged the fact that this was probably one workflow that synthetic biologists might be working on within what they do. And the great thing is with ExpressPlex being so quick, there would be plenty of time to do other things on the platform as well. So it was a tool that we could supply that could be used for many things and this workflow being one of them. Curtis can explain a little bit more about the specific ExpressPlex workflow.
Improving the Status Quo with ExpressPlex
Curtis: Thanks, Paul. ExpressPlex has been really exciting for us to release. It really was designed for the synthetic biology world where there’s a lot of plasmids and amplicons, but what I want to do is first walk through what this workflow looks like, and it’ll be very obvious why something like this is easy to automate as well.
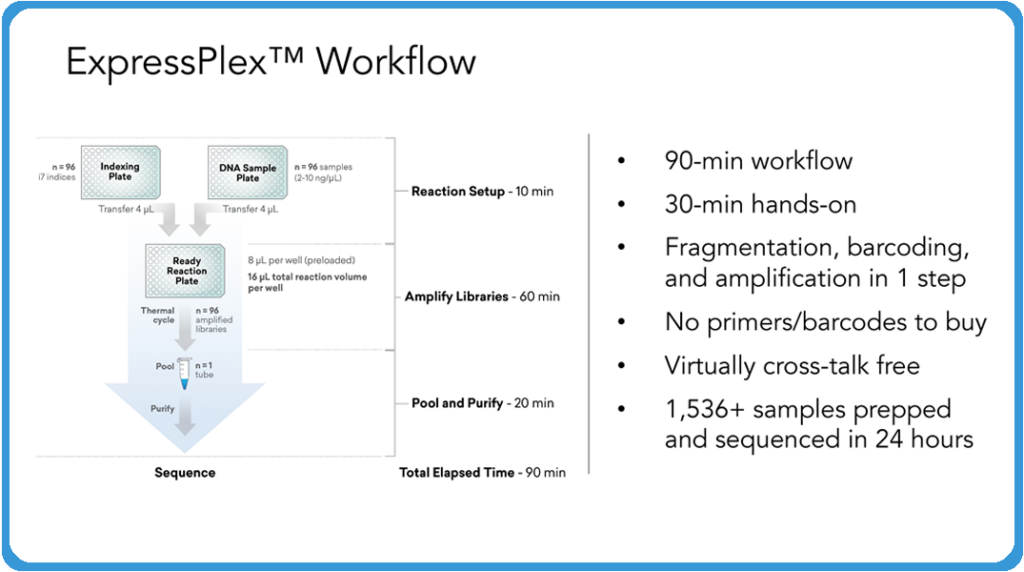
What you have with ExpressPlex is a system where you have two pipetting steps to begin with to really set up your system. You start with an indexing plate that has our tagging reagents on them and a reaction mix plate, or what we call a ready reaction mix. This is basically a master mix. And then you supply a plate of your DNA samples. The process is simply transferring 4 microliters of your indexing reagent into your ready reaction mix and 4 microliters of your DNA into that same ready reaction mix plate, then sealing that and putting that on the thermocycler.
After the thermocycling program is done, then you pool all the samples into a single tube for that plate and then do a single paramagnetic bead cleanup. Really what this comes down to is this is approximately a 90-minute workflow from start to finish, including the cleanup and everything else. There’s an extra 30 minutes roughly if you were to do this manually by hand. And that’s in the magnetic bead cleanup at the end, for the most part. What’s really exciting about this system and what makes it different from anything else is that you’re doing the fragmentation, the barcoding, the amplification – everything, all in one step.
Unlike some other tagmentation-, fragmentation-based systems, you don’t have to go out and buy your primers, nor do you have to buy your separate barcodes. Everything is built into the initial tagging step. So when the tagging happens, you are physically adding every bit of the Illumina compatible adapter from start to finish at the point of tagging and fragmentation. This creates a system where you really don’t have any chance for crosstalk between your samples because everything’s going through this system individually until the point of pooling and cleanup. We’re doing this in 96-well plates now and expanding this to 384 and other versions in the future as well. In the setup we’re describing, it offers the chance to run through as many as 1,536 samples, including the sequencing, in a 24-hour turnaround, which is really exciting for customers.
Enhanced Normalization
Curtis: Now, one of the really cool features of the ExpressPlex Kit is the normalization aspect of the system. And what I mean by normalization here is that we ask our customers, “How variable are your samples coming in?” This system is tolerant of really a tenfold input. We shoot for four to 40 nanograms. We like it in eight to 40 nanograms full input, but we’ve seen a tenfold input range works really well. And if you know roughly what your samples are coming out of your extraction or rolling circle amplification or however else you’re getting your DNA from, say your plasmids, just do a simple global dilution across that plate and you don’t have to go through and individually normalize your samples or anything like that to get it into range.
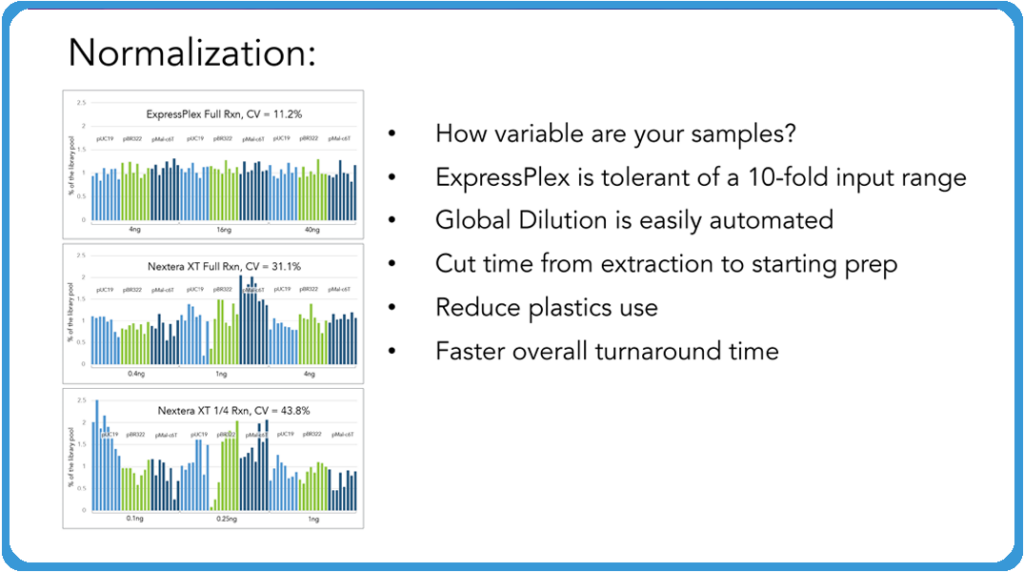
What we’re showing above is a comparison of ExpressPlex versus Nextera XT, which is one of the more common systems that people would use for this. You can see the variability that you can get in your read count levels across those samples when you are individually normalizing them. And the more normal your samples are coming out of that is something that allows you to save time, save sequencing costs, and as you’ll see in the image below, the ability to actually assemble your plasmids very quickly.
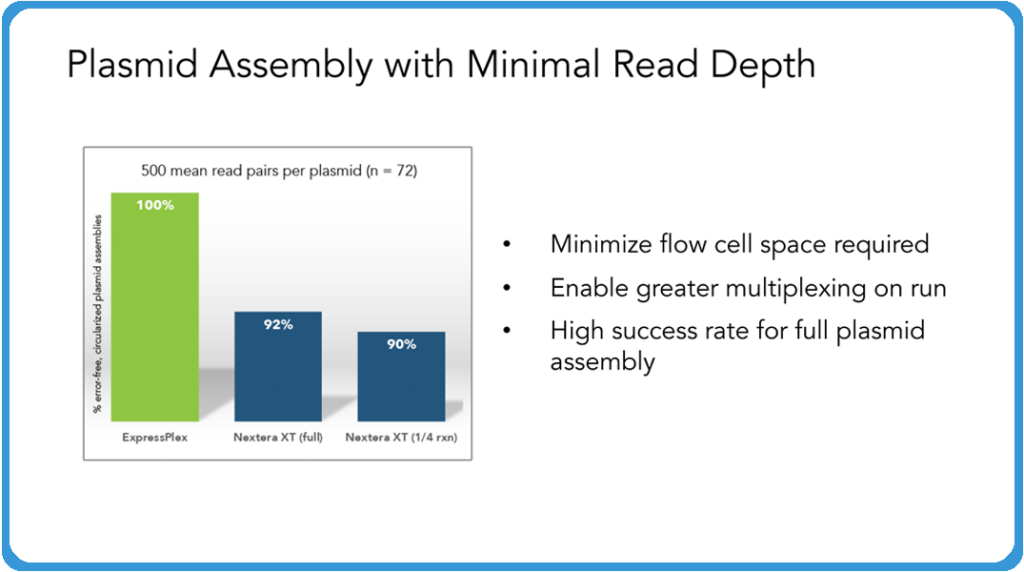
So with that in mind, as we start to think about plasmid assembly, what we’ve found is that even at fairly low read depths here, we’re describing just 500 mean read pairs per plasmid, we’re able to get 100% error-free, circlized, plasmid assemblies. And we see this routinely where you’re not necessarily getting that at these levels, you have to go up to higher numbers of read depth in order to get those full assembly. So one of the thoughts here is that higher success rates as well as the potential to actually multiplex more samples (even non plasmid samples) allows you to maximize the use of your flow cell there, resulting in cutting down your sequencing costs overall. And because of this, we’re expanding this product to 6,144 barcodes so you’ll be able to do even more samples throughout this system.
So as you think about this workflow overall, and consider how simple it is, you can understand how it made a lot of sense to partner with someone like SPT in order to automate this. We’ve actually done the ExpressPlex workflow on the apricot in-house and also in a miniaturized version with the mosquito. But we focused on the firefly here for the throughput aspects of it. Paul can talk about how this is set up on the firefly and what the workflow looks like there.
Automating ExpressPlex on firefly
Paul: Thanks, Curtis. Well, the key word here is simplicity. It really is a nice, simple workflow to automate.

So as you can see from the image above, these are just a few screenshots from the software. The key point here is the whole protocol is shown on the top screen. It’s a very visual UI, but there’s only nine steps in it. And in terms of what we need to load onto the deck, there’s just two tick boxes and three plates within the UI, there’s a really nice loading guide which walks the user through setup. This really could be used by somebody who’s never used automation before. They could follow the guide, load it and go. It’s really that straightforward. From our experience, it doesn’t get more simple than this. ExpressPlex is really a beautiful kit to automate. And I think at that point, it’s good to just have a look at the results.
A Performance Comparison
Curtis: As part of this validation between the two companies, we sent over everything, including the reagents and the DNA samples, and then we had the SPT team run multiple plates through this, and then we compared directly to a manual plate that we prepared in-house. Then we took the plates prepared with the automation system and the manual prepared plates and ran them all together on the sequencer. The results are just beautiful!
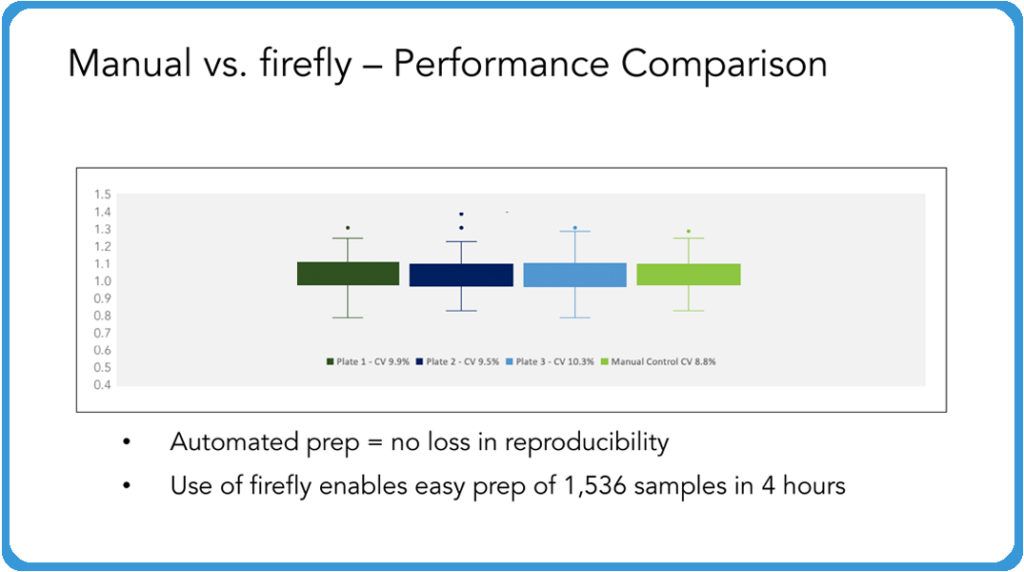
You could lay these on top of each other, absolutely no loss of reproducibility. And this is exactly what you want your automation system to do is to be able to just run this time after time after time in such a way that you’re allowing your techs to do other things or to be able to feed this system in such a fashion that you can turn around a lot of samples very, very quickly, without any concern about the reproducibility of these samples going forward. And so while I don’t show assemblies or anything here, really what we’re looking at is reproducibility and we couldn’t be more pleased with how tight these are in comparison across the board.
Paul: Yeah, it was great to see such great results. And as I say, it just went so smoothly in terms of automating. I think one thing that you’ve touched upon throughout this and where we started out at the beginning was the need for high throughput. And I think what we see from this point is that there’s the ability to push this a lot further in terms of the throughput. And you mentioned I think 384-well plates and greater numbers of indexes, so I think there’s plenty of scope to actually push this even further than we have already.
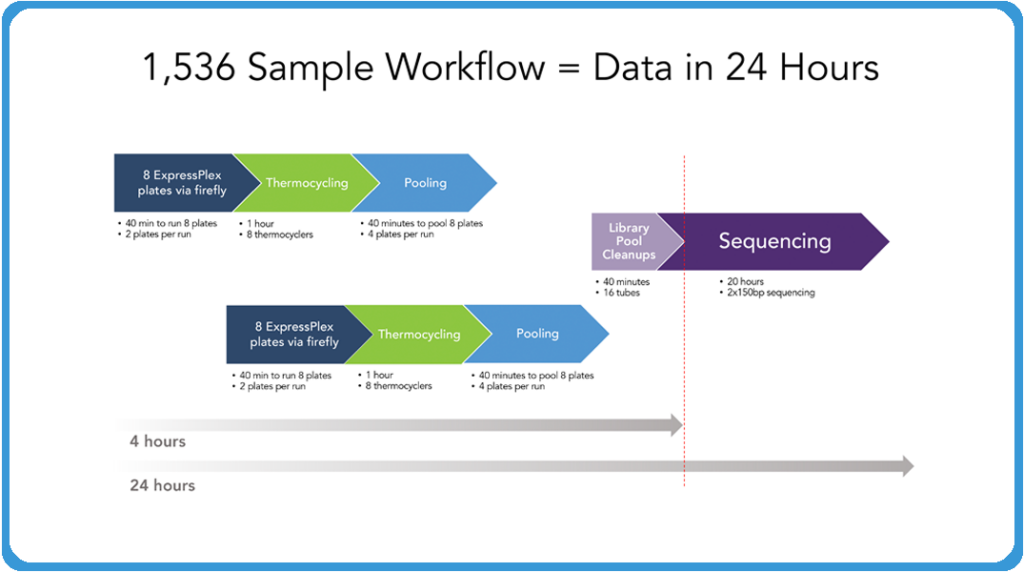
The image above demonstrates what it takes now to do 1,536 samples and have that, from the point of extracted DNA to the point of actual sequence data, in 24 hours. I should note that everything we’ve shown up to this point has been doing one plate at a time. It’s very easy to run two plates at a time for the physical setup of the libraries to put them on the thermocyclers.
It only takes a few minutes to do each of those plates. In 40 minutes, you can have eight plates set up and put those on your thermocyclers. While those are thermocycling, you can have another set of eight plates running in order to generate 16 total plates, which is your 1,536 samples. At the point at which that thermocycling is done, pull those plates back, then turn around your second set and put those on the thermocyclers. You can be pooling your original eight and you can do those four plates at a time. As you see, it takes very little to pool those plates. And then at the end of that you’ve got 16 individual tubes of libraries that we’ve typically been doing that library pool or cleanup individually by hand because it really doesn’t take much to do that. But obviously automation companies are very capable of doing that type of thing as well, so it wouldn’t take much to continue to do that in the end.
And in four hours you have your 1,536 samples set up, at which point you put it on your sequencer and run it overnight. It’s approximately a 19 to 20 hour run on a MiSeq for a two by 150 run. And in the morning you’re coming in and by midday you’ve got your data back at which point you can be starting the next set. So it’s something that one individual with the firefly can be running through a lot of samples very quickly to generate that data so that the companies can be focusing on their discovery and understanding where they want to go. And as I said, we’re expanding the barcode set to 6,144 barcodes. Our ExpressPlex HT 384-well ultra-high throughput version of this product is available as early access now. So as your throughput scales up, between the two companies we can scale with you in order to really help you get through massive amounts of discovery pretty quickly.
Closing Remarks
Curtis: Whether this is something you do every day, 365 days a year, or a big project push where you need to get through a lot of samples very quickly so that you can then sit and analyze your data and decide what’s next, a system like this works well. It means you’re not spending all of your time on the bench, you’re spending your time focusing on the discovery and getting to that end result.
Paul: Yeah. Thanks, Curtis. I think the two main takeaways are that we’re providing new tools here with this combination for a much faster turnaround of those critical experiments. And I think one of the things you stressed time and time again, is that it’s about enabling scientists to focus on discovery and actually not just standing at the bench pipetting.
