
Introduction
There’s no question that the world of biotech has embraced next-generation sequencing (NGS). Despite driving developments and discoveries among many applications, NGS continues to be a complex technique that has its share of technical difficulties.1
In fact, NGS library prep in particular, is often considered one of the more challenging steps of NGS, as it is a “significant source of inefficiencies that could hinder research and stifle progress by wasting resources and increasing costs.” 1
So how can scientists avoid some of the challenges during their NGS library prep? One way is by making sure NGS library QC is integrated into their process and done correctly.
Two key steps in NGS library QC are normalization of library concentration and control of library size. Library concentration is often measured using qPCR or fluorometric assays for concentration. Performing this step on multiple samples typically involves time-consuming steps to set up assays that measure concentration, with multiple replicates per sample. The accurate measure of functional library concentration is also dependent on measuring the size distribution of library fragments in the samples.
These steps are important because if the concentration of the library is not correctly estimated several consequences can occur:
- If the library is pooled with other libraries for a multiplexed NGS run, the amount of data from each sample can deviate from the intended design of the experiment.
- If multiple libraries that have been misquantified are pooled together for a multiplexed NGS run, the concentration of the entire pool may not be calibrated accurately for loading onto the sequencing instrument, leading to over or under-loading, thus impacting cluster density, data yield and quality.
A low quality library can skew NGS data, not only affecting the reliability of reads, but also causing reproducibility issues.2
Top 3 Steps to Take for NGS Library QC3
There are three main QC steps researchers can perform prior to their NGS library prep:
- Nucleic Acid Quantitation: It’s vital to evaluate the concentration of nucleic acid samples to determine fit and amount of nucleic acid available for your process.
- Nucleic acid purity: Contamination of nucleic acid samples is always a threat in your process. These impurities can decrease the sensitivity and efficiency of your downstream steps.
- Nucleic acid integrity (size distribution): In conjunction with quantity and purity, size distribution is a critical QC parameter because it provides additional insights into sample quality.
Making Library QC Simpler
Using QC procedures to obtain an accurate measure of library concentration allows you to normalize your library, while achieving optimal yield and data quality.
For many conventional library preps, precise control of the quality and concentration of input DNA will help minimize the variability of library generation and make the process of library QC simpler.
Another approach is for the library prep process itself to control for sample variability – effectively making library QC part of library prep. A number of newer library prep products on the market achieve this in various ways.
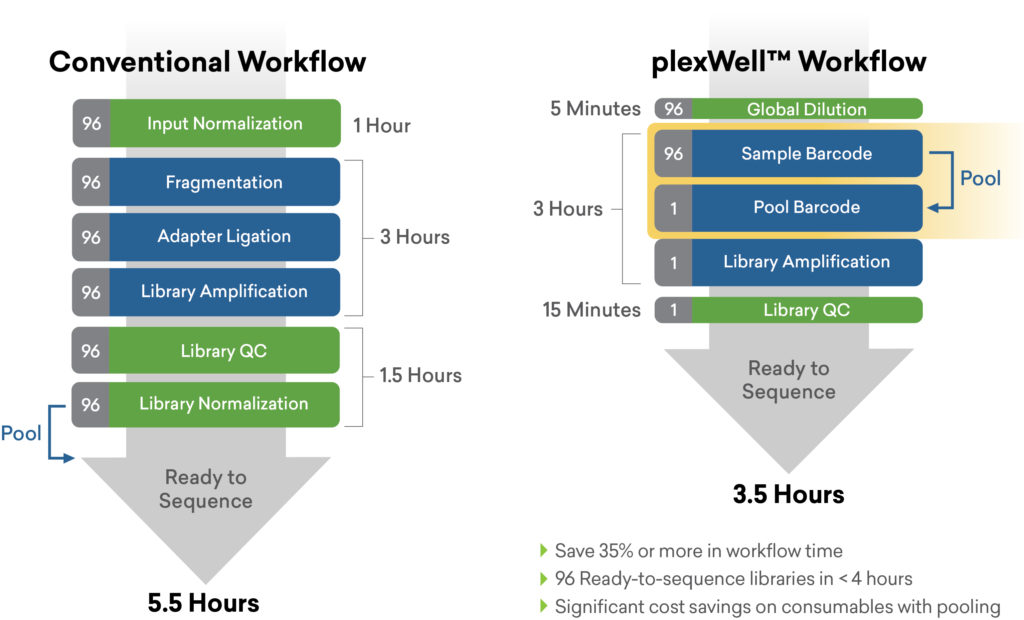
Utilizing a kit such as seqWell’s plexWell™ solution for preparing NGS libraries from hundreds to thousands of samples can help. The generation of library fragments is performed in a single pool format after a single tagging step, which has the combined benefits of normalizing for input DNA variation as well as sample-to-sample variation that can occur when libraries are prepared individually.4 This reduces associated labor, consumable and library QC costs by more than 10-fold. The simple and scalable plexWell workflow provides a greater consistency across samples and fewer handling steps downstream.4
Conclusion
In order for NGS to be successful, it’s critical to implement effective library QC procedures into your process. More importantly, it must be done correctly. For many conventional library preps, precise control of the quality and concentration of input DNA may help reduce library generation variability and simplify the library QC process. Using an effective technology such as seqWell’s plexWell library prep kits, is one way to achieve that.
References
- https://www.tecan.com/blog/the-benefits-of-optimizing-ngs-library-qc
- https://www.biocompare.com/Editorial-Articles/579849-NGS-Library-QC-and-Quantitation/
- https://blog.genohub.com/2017/09/15/sample-qc-steps-prior-to-library-preparation-for-ngs/
- http://seqwell.com/wp-content/uploads/2019/06/TDS_plexWell_v20181106.pdf
