Normalizing UDI Library Construction Poster Preview
Most available unique dual indexing (UDI) methods incorporate indexing into the library PCR step, which can be labor- and cost-intensive as libraries must be amplified, purified, quantified, and normalized individually prior to pooling for sequencing. In the purePlexTM DNA Library Preparation workflow, auto-normalization is achieved through sequential transposition events of full-length indexed adapters in the presence of a novel normalization reagent. This approach allows incorporation of unique dual indexing and permits pooling of samples immediately following the tagging steps such that purification and amplification of fragments occur after pooling, reducing the QC and labor costs over traditional UDI workflows.
A distinct advantage of the purePlex DNA library preparation workflow is the elimination of individual sample and library normalization. Herein we demonstrate the auto normalization of the purePlex DNA library preparation method in terms of reads per sample, median insert, and performance across bacterial genomes with GC content ranging from 29-69%. In contrast to other transposase mediated UDI library prep kits, such as Nextera XT DNA Library Preparation kit, the insert size is preserved across all samples without the need for careful normalization of the starting DNA and regardless of GC content.
Download the poster to read more.
Complete the form to download the poster.
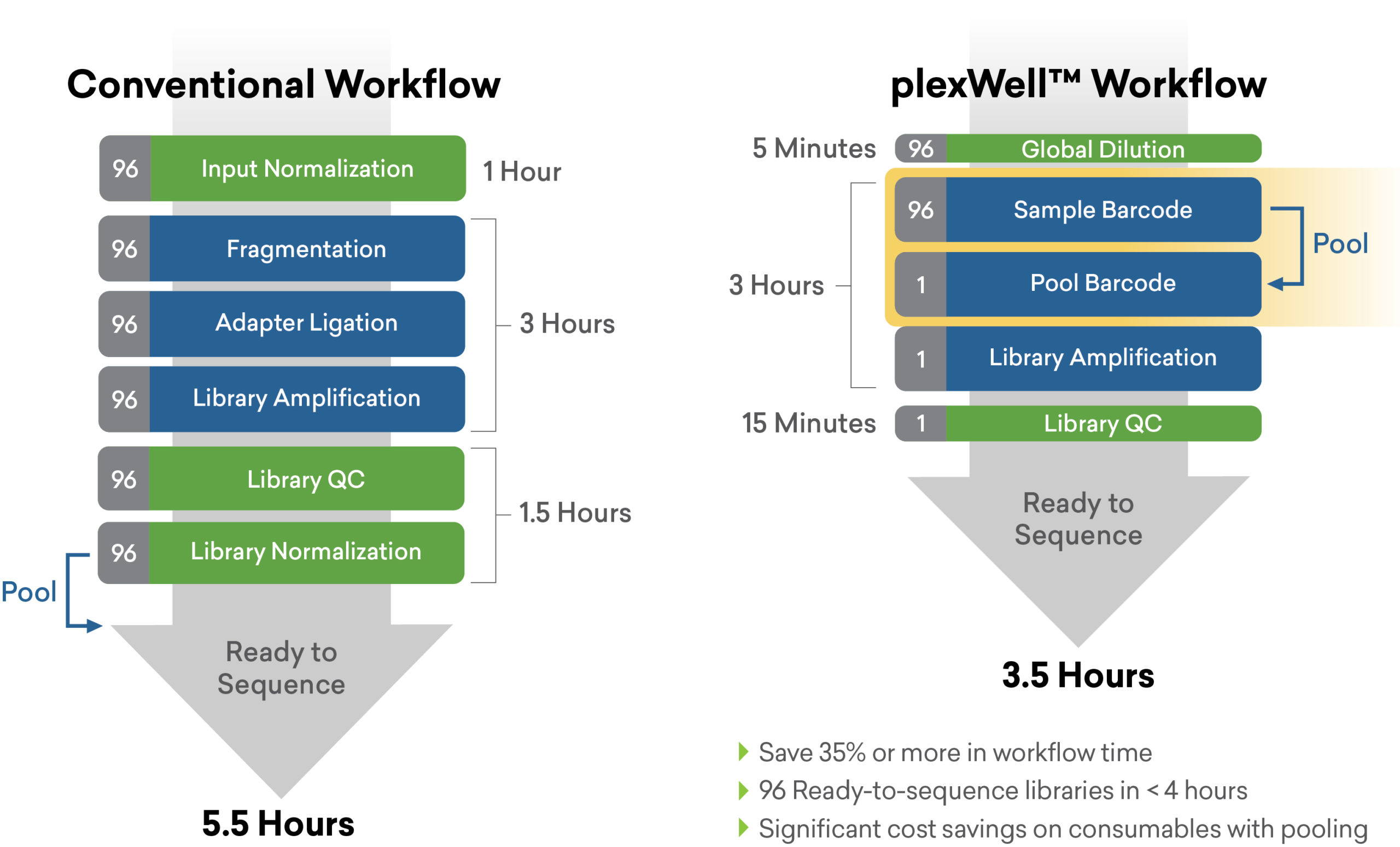
True multiplexing > Built-in normalization > Simplify library prep with plexWell
The plexWell difference
seqWell’s workflow and reagent engineering approach helps researchers harness the capacity of modern sequencing instruments to accelerate their research at scale.