MosaiX™ Library Prep Kit
Combine the workflow simplicity of tagmentation, the reduced insertion bias of TnX, & the performance benefits of ligation and you’ve got MosaiX Library Prep using seqWell’s directional tagmentation for population genomics….complexity made simple.
- TnX, next-generation transposase
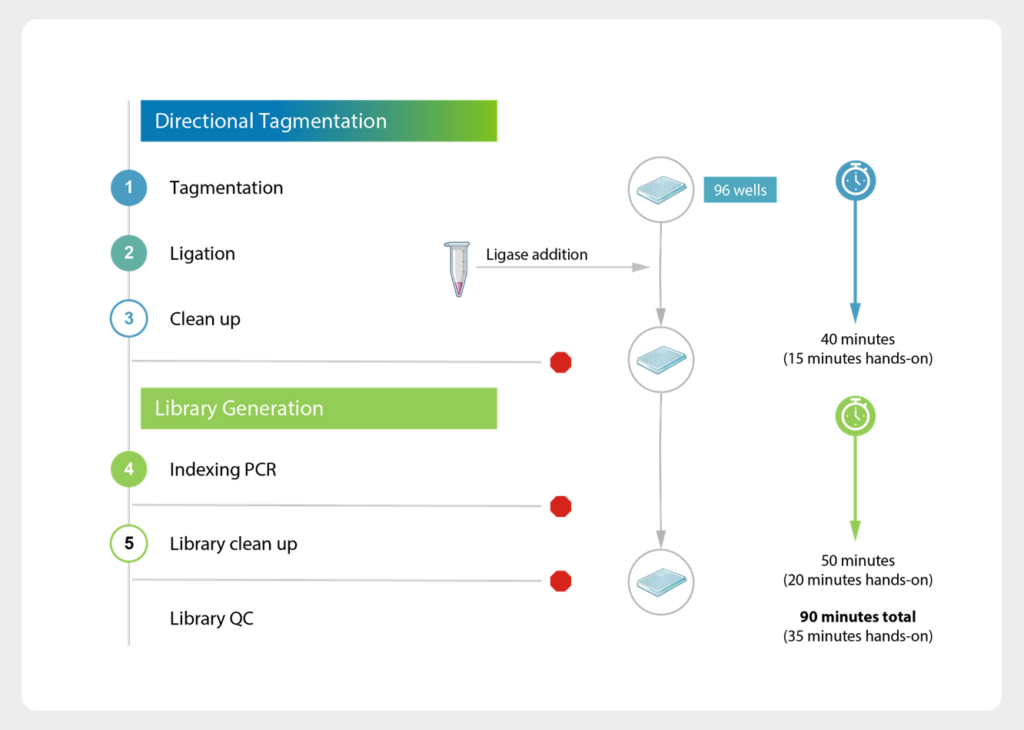
- Simple 90-minute workflow (35 minutes or less hands-on)
- Produces high complexity libraries
- Uniform sequence coverage
- Low duplication rates
seqWell’s Directional Tagmentation ...complexity made simple
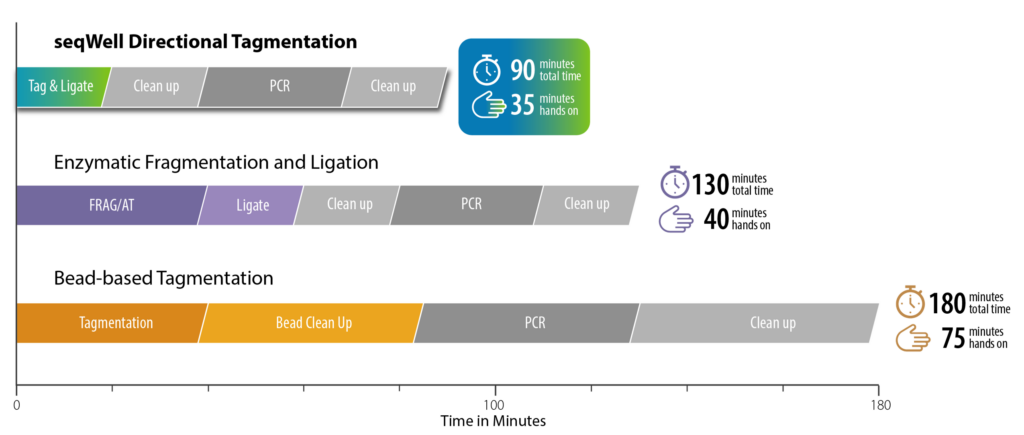
seqWell Directional Tagmentation
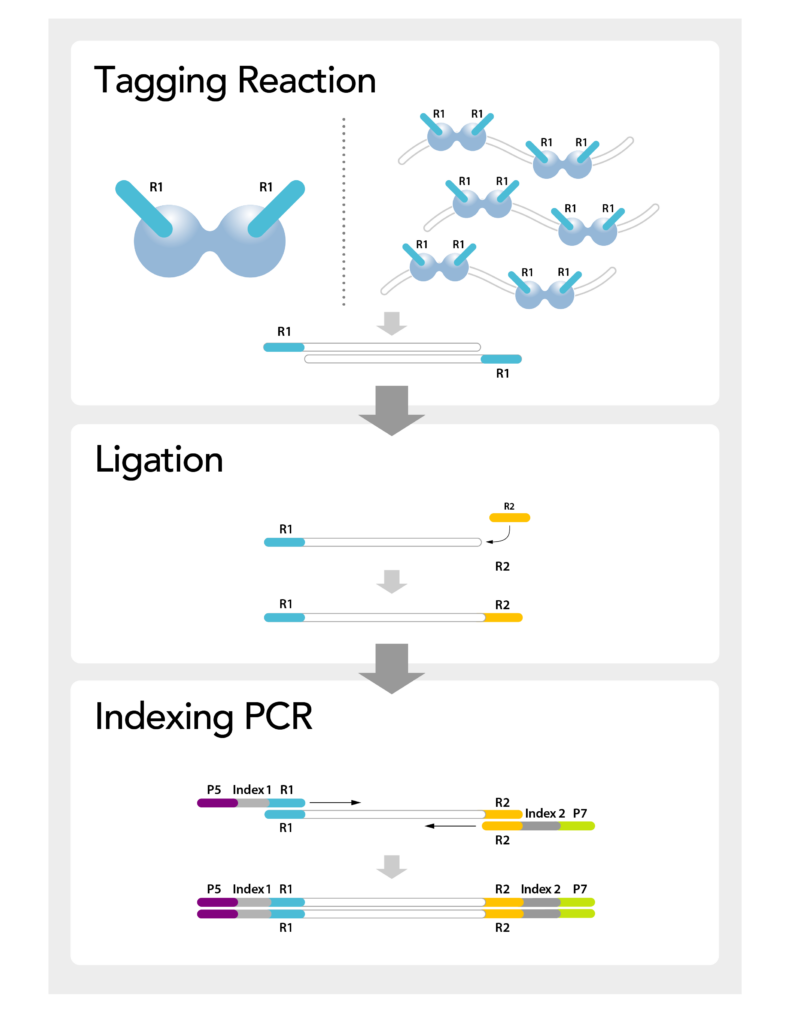
MosaiX 90-minute Workflow
“I’ve been really impressed with the outstanding performance of seqWell’s MosaiX library preparation kit. Directional tagging, driven by their TnX transposase, reliably resulted in libraries with high complexity and uniform coverage that provided for more precise sequencing data.”
– Massimo Delledonne, Professor of Genetics at University of Verona, Italy
The TnX Difference
Reduced insertion site bias
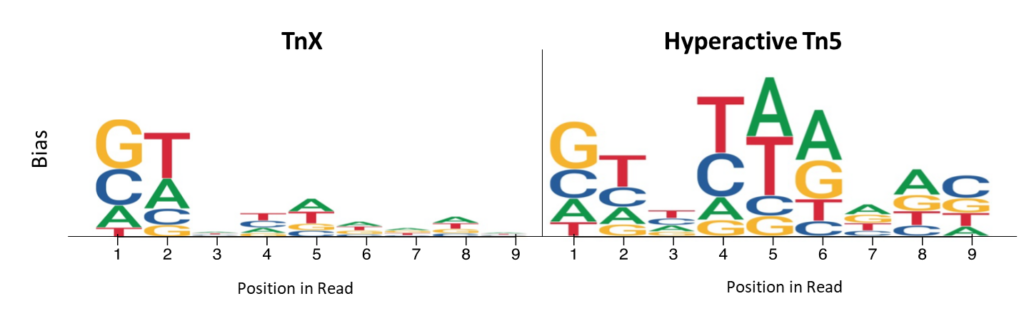
Read start site insertion bias was measured by examining the frequency of bases in the first 9 bases of each read. Positions with higher per-base nucleotide bias are represented by heights for hyperactive Tn5 and TnX, and illustrate the reduced bias of TnX.
The Performance of Ligation, with the Ease of Tagmentation
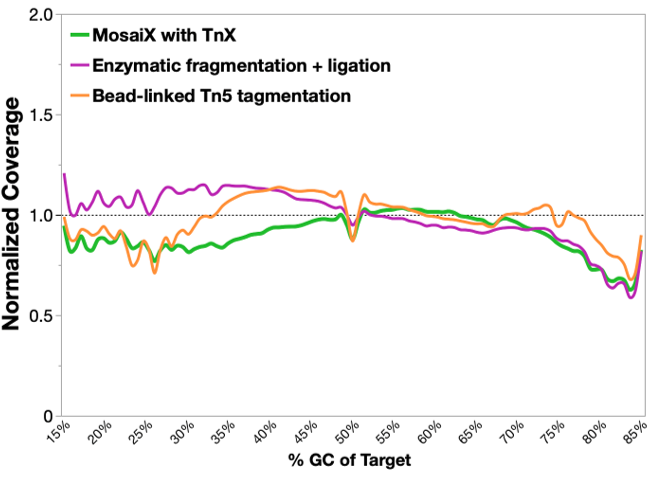
Whole Exome Sequencing
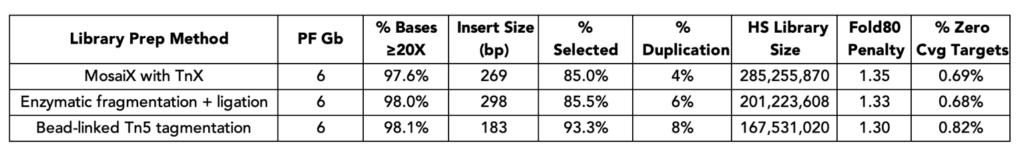
Exome metrics at 6 Gb for libraries generated using MosaiX Library Prep were extremely comparable to libraries generated using enzymatic fragmentation followed by ligation. Additionally, MosaiX Library Prep outperformed bead-linked Tn5 transposition in terms of duplication rate, library complexity (HS Library Size), and % of Zero Coverage Targets – highlighting the TnX performance difference!
Method: 50 ng of NA12878 DNA (Genome in a Bottle) was used for all and libraries were prepared following manufacturers’ user guides. Libraries were then captured using Twist’s Exome 2.0 panel and standard workflow. Sequencing was performed on NextSeq 2000, down-sampled to 6 Gb each, then aligned to Twist exome capture targets on hg38.
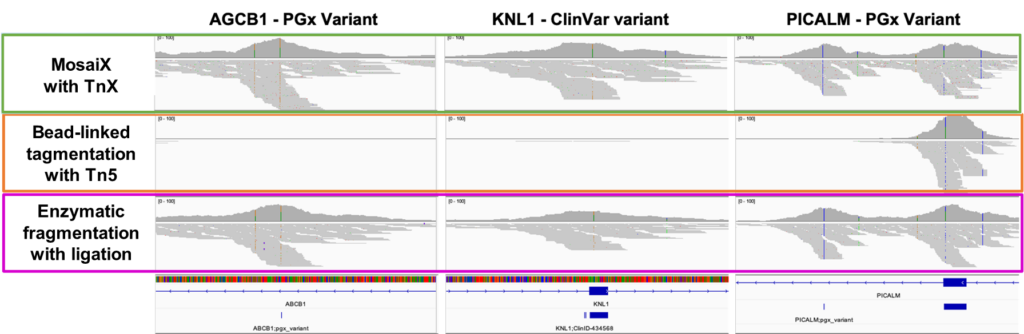
TnX finds those missing exome targets!
- Clinically relevant targets can be missed when using other library preparation methods such as bead-linked tagmentation using Tn5
- Reduced TnX insertion bias and higher molecular complexity of MosaiX libraries can access difficult regions of the genome that Tn5 transposase cannot.
Whole Genome Sequencing
MosaiX libraries achieve higher coverage compared to libraries prepared with bead-linked Tn5 transposition with the same total PF Gb. Duplication rate is lower and estimated library size is higher for MosaiX, indicating a more complex whole genome library.
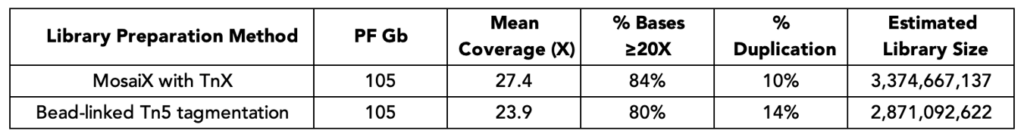
Method: 50 ng of NA12878 DNA (Genome in a Bottle) was used in both and libraries were prepared following manufacturers’ user guides. Sequencing was performed on a lane of a NovaSeq X+ 25B flow cell, down-sampled to 105 Gb each, then aligned to hg38.

POSTER: MosaiX- High-Performance DNA Library Prep with Directional Tagmentation Powered by TnX
View Now
MosaiX Specifications
| MosaiX Specifications | |
| Primary Applications |
|
| Transposase | TnX – Next generation engineered transposase |
| Sample Input types | Genomic DNA |
| DNA Input Range | 1 – 50 ng* |
| Buffer Compatibility | DNA diluted in: 10 mM Tris-HCl, 1X TE, Low TE, or Water |
| Total Library Prep Time | 90 minutes (35 minutes hands-on time) |
| Mean Output Fragment Size**
Fragment sizes as measured by TapeStation (including adapters) |
WGS recommended 0.7x bead clean up protocol: 850 bp ± 15%
Target capture recommended 0.8x bead clean up protocol: 700 bp ± 15% |
| Indexing Strategy |
|
| Supported Paired Reads (clusters/sample) | ≥400 million |
| Reactions per Kit | 24 or 96 |
| Sequencer Compatibility |
|
* < 5 ng may require optimization of adapter concentration and PCR cycles
** Fragment size optimized for NovaSeq X Plus
Early Access MosaiX Library Prep Kit Includes:
- TnX Read 1 Tagging Reagent
- 5X Reaction Buffer
- Tagmentation Enhancer
- Read 2 Adapter
- DNA Ligase
- 2X Amplification Ready Mix
- MAGwise Paramagnetic Beads
- Diluent