Introduction
People ask me on occasion where the idea to create seqWell came from. My response starts with my own experiences and challenges faced as a scientist and developer – a lot of time spent in a lab, using molecular biology and pipettes to solve different research problems with sequencing data, and even more time spent with a computer trying to analyze and interpret that data. Those experiences made me strongly believe there had to be a better way to use sequencers to create high-quality data more quickly and easily.
Thinking about and discussing the problems and challenges scientists face with next generation sequencing is the catalyst for developing ideas and advances that provide improved tools for life science. But sometimes, those advancements can upset the balance of, or need for, something else. The evolution of sequencing is a perfect example.
In this blog, I’m going to explain some of the challenges that modern sequencing technology creates, and why NGS multiplexing has emerged as a core and crucial methodology in genomics.
The Evolution of Sequencing
Anyone doing sequencing understands that the cost has gone down significantly over the years due to the increased scale of sequencers.
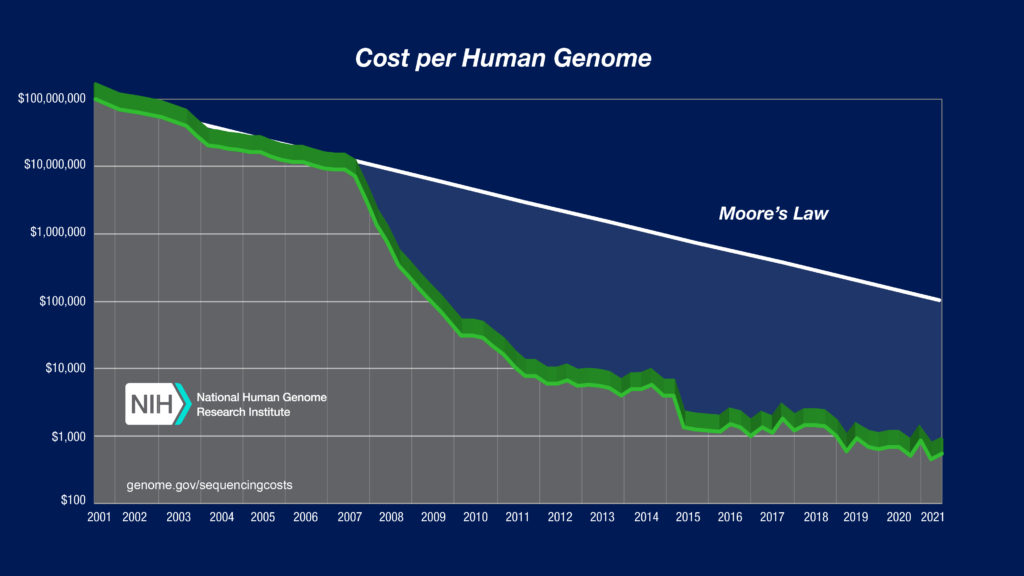
The image below shows the evolution of sequencing technology over the last 20 years.
This chart, updated regularly by the National Human Genome Research Institute, tracks how the exponential drop in the cost of sequencing has led to a current ability to generate the data needed to sequence a human genome for less than US$1000. This is an astounding record of technological achievement, and yet only part of the remarkable story of sequencing technology over the last two decades.
Missing from this description are structural changes in how sequencing is done that has accompanied this progress – changes that now permeate nearly every area of life science research.
The Scale Revolution
Much like how Moore’s Law is underpinned by constant improvement in the number of transistors placed in a given area on a computer chip, sequencing cost has been driven by being able to generate more reads in a single sequencing run. The typical benchtop sequencer today can produce more data in a day than the entire world produced in a single year twenty years ago.
On the flip side, this trend means that in order to take advantage of the low cost of sequencing that this high scale affords, sequencers and projects are run at a larger scale – in other words, more samples and more data per sample.
Increased Sequencer Scale has Created New Bottlenecks
Because sequencing capacity has increased so rapidly, for the vast majority of needs and applications, the cost of actually acquiring raw sequencing data is now much less than other costs associated with sequencing and analyzing a sample. These other costs – sample isolation, purification, library generation, data analysis and storage – are now larger than they have ever been before relative to the cost of generating sequencing data, and this means that new bottlenecks have emerged.
Steps that were negligible in a lower-throughput era are now often prohibitive to efficiently utilize the throughput of large sequencers such as the Illumina NovaSeq.
Multiplexing Tools Unlock the Power of Sequencers
The key innovation that sits at the intersection of the increased scale and new bottlenecks created by modern sequencers is multiplexing. Multiplexed sequencing assays allow the bandwidth of sequencing instruments to be spread over larger numbers of sample types ranging widely from cells and transcriptomes to genomes and epigenomes. By dividing the capacity of a sequencing run over a large number of samples, the cost-per-sample decreases while the number of samples that can be simultaneously analyzed is greater.
The trend toward increased multiplexing has exploded in the form of sequencing workflows that can be used. An example of this is to analyze many thousands of cells, or sequence thousands of viral samples such as SARS-CoV-2, in a single run.
Summary of Observations
In summary, multiplexing is an incredibly powerful concept that is now the workhorse for an expanding number of sequencing applications. Multiplexing is like the leveler between the cheaper sequencing and the bottlenecks downstream. More NGS tools and products need to be developed to improve multiplexing, harness the capabilities of modern sequencing instruments, and to address the bottlenecks – and that’s what we’re focused on at seqWell.
About the Author
Joseph C. Mellor, PhD, is Co-founder and Chief Scientific Officer of seqWell Inc. Joe earned a Bachelor’s of Science degree with honors in Biochemistry and Chemistry from the University of Chicago, followed by a PhD in Bioinformatics from Boston University.
Prior to co-founding seqWell Inc. in 2014, Joe was an interdisciplinary scientist working at the intersection of computational biology and next-generation sequencing at institutions such as Harvard Medical School and the University of Toronto. Joe is an expert in all phases of next-generation sequencing (NGS) technologies, with an extensive publication record and history of productivity in fields ranging from molecular biology to bioinformatics. Joe brings a diverse set of skills and leadership experience to seqWell, and is co-inventor of seqWell’s core, proprietary NGS technologies .