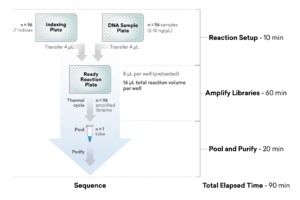
Go from extracted sample to libraries on sequencer in < 1/2 a day
Save time & cost with true multiplexed NGS libraries
seqWell Technology is Accelerating
NGS Research for Our Customers
ExpressPlex: A 90-Minute Library Prep Method
30 Minute Hands-On Time (for 96 Samples)
-
Construct multiplexed, balanced library pools in 90 minutes, suitable for same-day sequencing
-
Requires minimal liquid handling steps
-
Auto-normalizes insert size and read count
-
One pool cleanup

Powerful High-Throughput Sequencing
Unique transposase-based tagging method
ExpressPlex™ library preparation technology utilizes a unique, sequential, transposase-based tagging method to generate auto-normalized NGS libraries in true multiplexed fashion. This approach is particularly powerful in high-throughput sequencing of samples with relatively low sequence complexity (e.g., Plasmids, and small genomes), where it is challenging to balance library construction capacity and cost with available sequencing capacity.

A Unique Approach to Library Prep
Built-in Normalization = More Cost-effective Sequencing
The cost-effective, streamlined, and highly scalable workflow yields pools of normalized libraries with unique barcode combinations. Uniform read distributions and high data quality further reduces overall sequencing costs.
-
Only 3 pipette transfers
-
Flexible batch sizes, combinatorial indexing
-
Built-in read count and insert size normalization
-
Designed for support of automation and flexible reaction volumes
ExpressPlex is the latest of seqWell’s family of products, including:
Tn5 Transposase

The technology developed by seqWell permits scientists to create personalized reagents by designing and synthesizing them according to their research objectives. This feature allows scientists to optimize a reagent for a particular application, such as including Tn5 transposase and a unique payload suited to their research goals.
plexWell™ 96

plexWell™ 96 uses SeqWell’s proprietary plexWell™ technology, which involves tagging individual DNA molecules with unique barcodes that enable accurate and efficient sequencing of large numbers of samples in parallel.
plexWell™ 384

plexWell™ 384 supports a wide range of sequencing applications, including whole-genome sequencing, exome sequencing, targeted sequencing, and single-cell sequencing
Perfect for High-throughput Plasmid and Amplicon sequencing
High Success Rates of Plasmid Assembly
Given the robust read balance and lower number of reads required to generate correct assemblies, ExpressPlex users can multiplex higher numbers of samples on a single Illumina run or utilize a lower capacity flow cell, thereby reducing the cost of sequencing per plasmid.
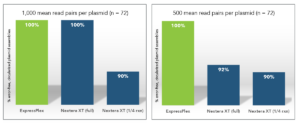
Libraries were prepared as described above, sequenced (MiSeq Nano; 2 x150) and de novo assembled using seqWell’s SNAP plasmid assembly pipeline. The sequencing run was downsampled to either 1,000 or 500 average read pairs per sample to identify error-free, circular plasmid assemblies. Even at only 500 average read pairs all ExpressPlex samples generated correct circularized assemblies. At the same number of reads, however, Nextera XT (full and ¼ reactions) failed 8-10% of assembly attempts.
Given the robust read balance and lower number of reads required to generate correct assemblies, ExpressPlex users can multiplex higher numbers of samples on a single Illumina run or utilize a lower capacity flow cell, thereby reducing the cost of sequencing per plasmid.
Confident Amplicon Sequencing
ExpressPlex’s workflow and 10bp barcodes provide >99% on-target rates.

20 uniquely mapping amplicons were generated from lambda DNA. The amplicons were used combinatorial as input into the ExpressPlex library prep such that each sample is comprised of 2 lambda amplicons and each well contains a unique amplicon combination. Sample input was maintained at 16 ng and 15 cycles of PCR were used during the library preparation. Following sequencing, demultiplexed reads were mapped to the lambda genome and marked as on-target (expected amplicon combination) or off-target for each well. All wells have an on-target rate >99% (above figure) with an average on-target rate of 99.XX%. Amplicon sequencing with EP enables efficient preparation of up to 1536 amplicons multiplexed on a single lane, enabling high-throughput applications without fear of index-hopping, or PCR chimeras.
ExpressPlex is also ideal for:
Labs transitioning away from Sanger method due to need to process at larger volumes
Viral detection/sequencing
Applications that focus on speed and volume vs. high-depth reads
Learn more about how ExpressPlex technology can maximize your sequencing efforts.
Benefits of ExpressPlex
ExpressPlex technology is an ultrafast, low touch, automatable library prep
Extremely simple kit (only 3 kit components)
Low touch single reaction for library generation and amplification
Flexible batch sizes (8 – 1536 samples)
Easiest library prep workflow (One-and-done)
Automation-friendly
Tunable library prep platform for a broad range of applications
Save over 50% of time by using ExpressPlex. Check out the complete workflow
Less Plastics – Help Save the Environment
Thanks to the highly efficient ExpressPlex workflow, you can use almost 80% less pipette tips and plastics to prepare your libraries.**
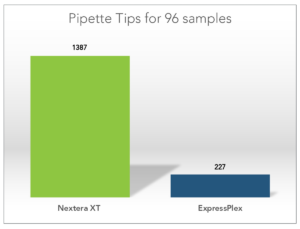
**calculations based on 96 samples prepared, comparing ExpressPlex to Nextera XT.
Complete the form to be contacted by a seqWell representative.
ExpressPlex = Better Performance
ExpressPlex demonstrates significantly higher levels of normalization compared to competitors, enabling a simplified workflow where individual normalization is no longer required to achieve more consistent read-depths across samples.
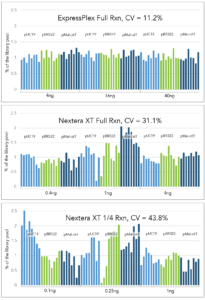
Libraries were prepared from three different reference plasmids of varying sizes across a 10-fold input range at full reaction volume for ExpressPlex (16 µL) and Nextera XT (50 µL), as well as at ¼ miniaturized reaction volume for Nextera XT.
For each method, inputs ranged from 4-40ng for ExpressPlex (standard input is 16ng) and 0.4 – 4ng for Nextera XT (standard input is 1ng). Inputs were scaled appropriately for 1/4 reactions.
Auto-Normalization of Insert Size
ExpressPlex generates highly consistent library insert sizes over a broad range of DNA inputs
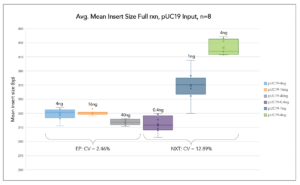
Libraries from pUC19 plasmid DNA were simultaneously prepared at 3 different input amounts (n=8 each input amount). ExpressPlex demonstrated greater consistency in average insert size across a 10-fold range.
Specifications
| Specs | Description |
|---|---|
| Primary Applications* | Plasmid and amplicon sequencing |
| Sample Types* | Plasmids, amplicons > 350bp |
| Reactions per Kit | 96 reactions
384 (4 different sets available) |
| DNA Input Recommended | 8 – 40ng |
| Total Library Prep Time (hands-on time) | 90 min
(30 min hands-on) |
| Indexing Method | Combinatorial Dual Indexing |
| Number of Unique Index Combinations | 1,536 |
| Supported Paired Reads (Clusters) per Sample | < 4 million (validated to date) |
| Output Fragment Range** | 400 – 1,200bp |
| Number of PCR Cycles | 12 cycles for plasmids 15 cycles for amplicons |
| Sequencer Compatibility | All Illumina sequencing platforms; Use with Element Biosciences AVITI™ or other sequencing platforms is possible with conversion kits for Illumina libraries |
*Other sample types are compatible. Contact seqWell’s application group for guidance.
**Fragment size will depend on magnetic bead cleanup ratios used.
ExpressPlex Library Prep Kit Includes:
Indexing Reaction Plate(s)
Ready Reaction Mix Plate(s)
MAGwise Paramagnetic Beads
Users do not need to supply polymerase, primers, or barcodes. These are already pre-mixed into the reagents supplied.












