PhiRx™: A Color-Balanced Spike-In Control for Improving Demultiplexing on XLEAP-SBS Illumina Sequencing Instruments
Color Balancing and Illumina Sequencing
Color balancing is a crucial aspect of sequencing-by-synthesis (SBS) technology on Illumina platforms. Illumina-based sequencers have historically utilized a range of different fluorometric color schemes: Early Illumina instruments (e.g., MiSeq and HiSeq era, 2007-2015) used four different color signals for each base; more recent instruments have utilized an evolving series of two-color encoding schemes, beginning with the release of the NextSeq 500’s red/green system in 2015.
In 2023, Illumina released the NovaSeq X Series instruments that introduced the new XLEAP-SBS chemistry, a chemistry that has since been implemented on an increasing number of current instrument platforms including the NextSeq 1000/2000 Sequencing Systems, and the newly launched MiSeq i100. In addition to improving sequencing chemistry for higher throughput and longer reads, the relatively new XLEAP-SBS technology has also introduced a new fluorescent color scheme. An overview of two-color schemes and differences on recent instruments is shown below:
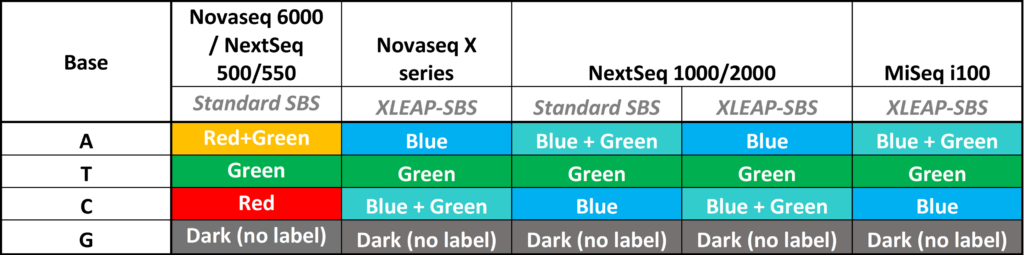
The Problem: Color Balancing and XLEAP-SBS Index Failures
A simple example of “failed” color balancing is when all of the sequences in a lane produce exactly the same signal (color), this would be more likely to occur where the index sequences on a library are very similar or do not produce a high enough diversity of color signals.
One of the more common pitfalls that users can encounter is that the nucleotide sequences of sample-specific barcodes do not always satisfy the optimal requirements to achieve balanced color signals when read by the sequencing instrument.
Despite its many benefits, XLEAP-SBS has presented users with challenges because previously well-performing index sequences (or combinations) can fail to satisfy the new requirements of updated color scheme chemistry.
Two examples of this are:
- Sequencing low-complexity libraries or Combinatorial Dual Indexed (CDI) libraries, the lack of base diversity during certain cycles can lead to severe color balancing issues.
- Indexing cycles lacking green signals cannot be correctly registered, leading to base-calling failures and a significantly higher percentage of NoCall.
These often result in a substantial increase in the number of undetermined reads and a higher rate of >1 mismatch reads, ultimately causing a significant loss of sequencing data.
Choosing Optimal Indexes for XLEAP-SBS
Selecting appropriate indexes is critical for achieving optimal sequencing performance with XLEAP-SBS.
Avoiding Low Diversity: A key principle is to ensure that each sequencing cycle includes a balanced representation of all four nucleotides (A, T, C, G). Low diversity can compromise color balancing and reduce accuracy. Illumina provides guidelines for pooling libraries to maintain nucleotide diversity, which is essential for optimal performance [2].
Illumina can only currently guarantee optimal index performance for the index sets they have validated and selected for their kits. Vendors that supply their own index combinations, that have previously been successful with Illumina sequencing, are currently struggling to find combinations that are routinely optimal for XLEAP-SBS.
PhiRx: A Color Balancing Index Control For Improved Performance on XLEAP-SBS Systems
To address the challenges of color balancing of different indexed libraries on XLEAP-SBS, including libraries made with seqWell library prep kits, we developed a new approach called PhiRx.
What is PhiRx?
PhiRx is an indexed control library derived from phiX174 genomic DNA that has been designed with a composition of bases at i5 and i7 index positions to achieve color balancing on Illumina XLEAP SBS systems. PhiRx is based on the idea that color balancing of index positions of sequencing libraries can be achieved effectively by adding a control library that artificially creates color diversity with a low-level spike-in of a carefully chosen mix of nucleotides at index positions.
PhiRx Indexed Control is specifically designed for use with NextSeq 2000 and NovaSeq X / X Plus systems, and has the following features and benefits:
- Optimizes Index Diversity: Provides a pre-configured mixture of nucleotides to support balanced color representation across all sequencing cycles, ensuring consistent compatibility with XLEAP-SBS.
- Serves as Seamless Spike-In Control: Functions effectively as a spike-in control library, similar to Illumina’s PhiX Sequencing Control v3, without requiring additional steps during pooling or sequencing.
- Corrects Index Color Balancing at Low Levels: Provides effective correction even at spike-in levels as low as ~5%.
- Improves Signal Uniformity and Data Quality: Enhances fluorescence signal stability while reducing noise and crosstalk, leading to increased accuracy for long reads and complex libraries.
- Helps recover data that would otherwise be lost as Undetermined Reads. In most real-world cases, the percentage of spike-in amount of PhiRx reads is substantially less than the total number of “extra” reads recovered.
An example of PhiRx in action is shown below, in Table 1. Here, a library with a suboptimal index configuration – a single i5 index with poor color-balancing properties – exhibits a high rate of demultiplexing failure (% Undetermined Reads) when loaded on a XLEAP-SBS NextSeq 2000 run. Spiking-in less than 5% of PhiRx Indexed Control reduces the rate of lost data from 53% to less than 2%.

We have also used PhiRx to address sequencing runs where they should have performed optimally, as they followed Illumina’s guidelines, but did not. In these runs we also see significant sequencing data recovery, similar to the above. PhiRx offers a promising, easy-to-use solution for researchers looking to maximize the performance of XLEAP-SBS on high-throughput Illumina platforms.
More Information about PhiRx Indexed Control
References
- Illumina. (n.d.). XLEAP-SBS chemistry enables Q40 and above data quality on NovaSeq X and NextSeq 1000/2000. Retrieved from (illumina.com)
- Illumina. (n.d.). Index color balancing for the NextSeq 1000/2000 system using standard SBS reagents. Retrieved from (knowledge.illumina.com)
- Illumina. (n.d.). Index color balancing for XLEAP SBS reagents on the NextSeq 1000/2000 and NovaSeq X/X Plus. Retrieved from (knowledge.illumina.com)

