Spend more time on data with faster library prep. Contact us today!

plexWell™ 96
- SKU: PW096
$1,975.00
plexWell™ 96 (PW096) Highlights:
- plexWell NGS multiplexed library generation kit for Illumina®
- Assay-ready 96-well fully-skirted low-profile PCR plate
- 96 unique barcode combinations
Description:
- Kit contains major reagents necessary to prepare libraries including Magwise™ paramagnetic beads (DNA polymerase not included)
- Efficient library prep for one 96-well plate inputs
- Normalizes input DNA over wide input range of 3-30 ng
- Volume-based pricing (save 30-50% on total lab costs)
Recommended Applications:
- Large scale full length viral surveillance
- Synthetic Construct Sequencing (Amplicons, Plasmids, BACs, etc)
- Microbiome screening
- Microbial whole genome sequencing
- scRNA-seq
- Low-depth whole genome/GBS
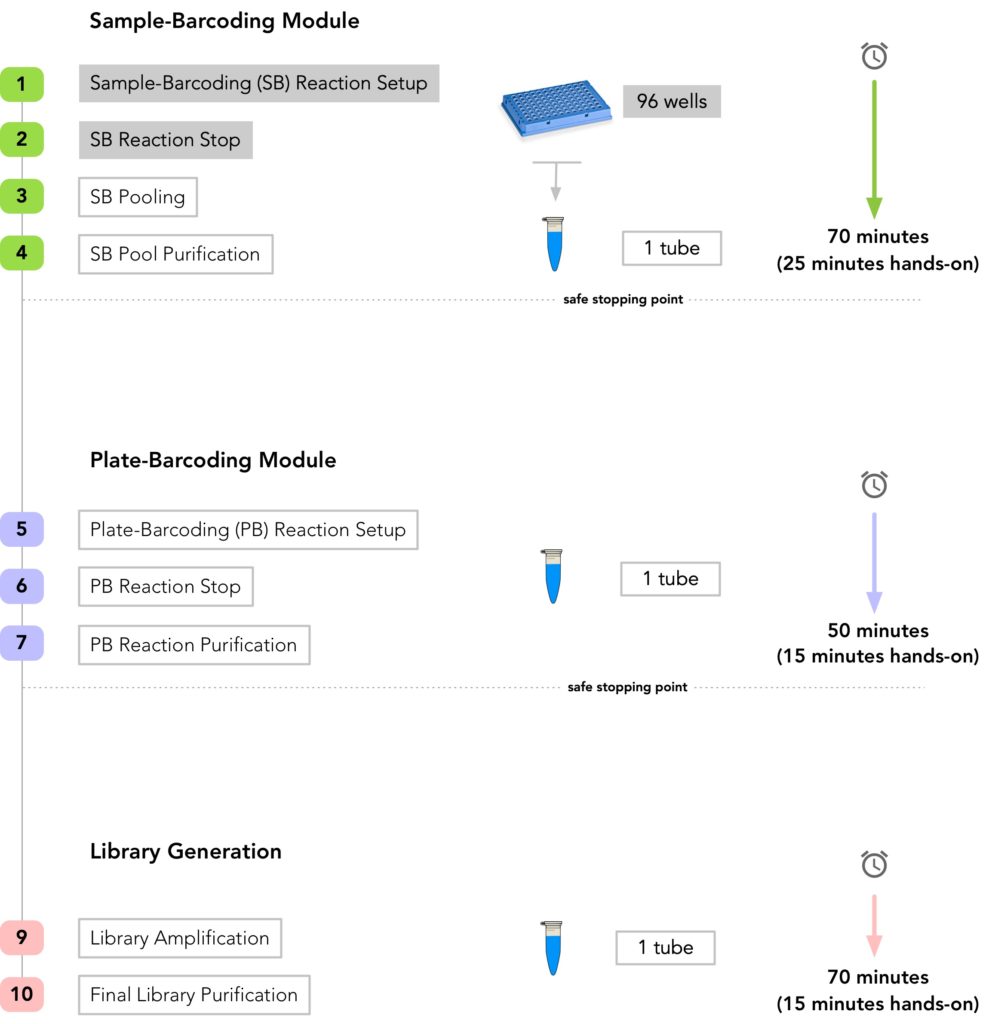
plexWell™ Library Prep Workflow:
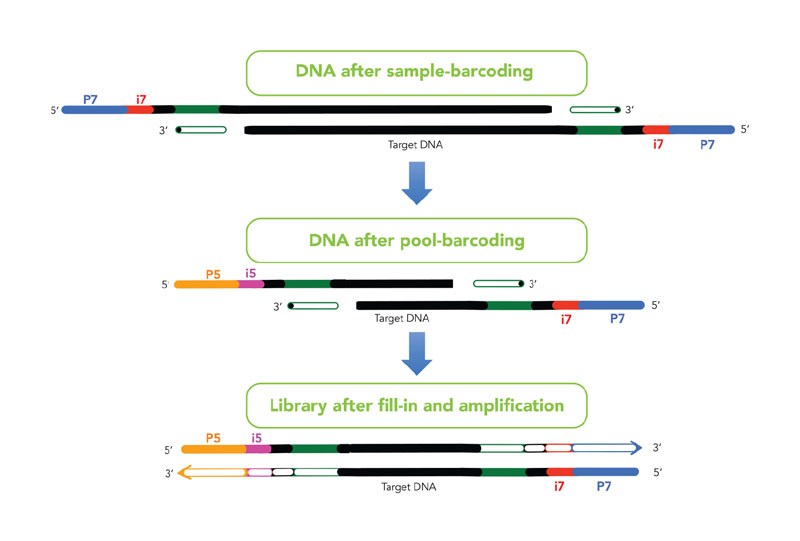
plexWell™ Library Prep Chemistry:
Specifications:
- Time-to-prepare a multiplexed library from 96 samples:
- Hands-on time: 1 hr
- Total elapsed time: 3 hrs
- Recommended input: 3-30 ng of genomic DNA per sample (n=96)
- Expected Results:
- Library Yield: 750 – 1500 fmoles of purified multiplexed library
- Library Diversity: <5 % dupe rate per 1M sequenced reads per sample
- Substantial normalization: Typically <2.5-fold range (min to max) in read count across all 96 samples
- Compatibility with Illumina sequencing systems:
- Compatible with Illumina MiSeq®, NextSeq®, HiSeq® and NovaSeq® instrument platforms
