Spend more time on data with faster library prep. Contact us today!
Discover an alternative to microarray-based genotyping that’s designed to scale.
Low-Pass WGS-Based Genotyping Application Note | Introduction
Advances in massively parallel sequencing and bioinformatics have enabled the routine use of whole-genome sequencing (WGS) in both research and clinical settings. While most applications require coverage depths in the range of 20X – 100X, low-pass or low-coverage (<1X) WGS is emerging as a powerful technique for detecting genome-wide genetic variation.
Download the Application Note to learn the wide-ranging benefits of high-throughput, low-pass whole-genome sequencing.
Suitable for a variety of applications
- Genome-wide association studies (GWAS)
- Animal and plant breeding
- Cell bank profiling
- Disease risk testing
Complete the form to be contacted by a seqWell representative.
True multiplexing > Built-in normalization > Simplify library prep with plexWell
The plexWell difference
- The plexWell™ LP 384 Library Preparation Kit and open source GLIMPSE pipeline offer an accessible, robust, and cost-effective pipeline for high-confidence genotyping based on lowpass WGS and imputation.
- 10 M read pairs per human genome (<1X coverage) is sufficient for precise and accurate genotyping, and offer significantly more information than microarrays.
- The plexWell LP 384 Library Preparation Kit requires less than one hour of hands-on time to generate two 48-library pools.
- The streamlined workflow and up to 2,304 barcode combinations facilitate implementation in high-throughput genotyping imputation pipelines.
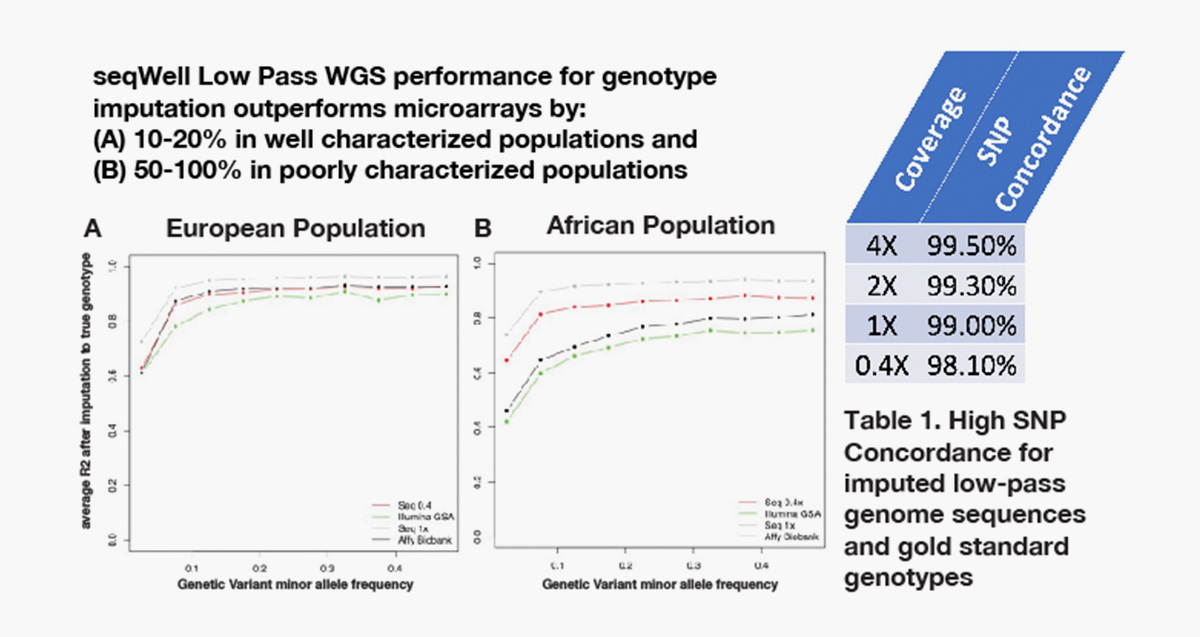
Gencove’s imputation SaaS in combination with seqWell’s low pass library preparation kit outperforms genotyping arrays.