Introduction
Ensuring the safety and efficacy of gene-editing-based therapies depends on rigorous gene editing quality control (QC), including both on- and off-target analysis. QC assays are critical for building confidence that engineered cells will perform as intended without introducing unintended risks.
High-quality assays must strike a delicate balance between delivering relevance that reflect true cellular outcomes, assay sensitivity to capture rare editing events, and reproducibility to enable trust across experiments and laboratories. At the same time, the field demands solutions that align with the scale of next-generation sequencing (NGS) projects to support large cohort and therapeutic studies.
The Power of Tagmentation: From CIRCLE-seq to CHANGE-seq
One promising trend driving the evolution of first-generation off-target assays to address scalability limitations is the incorporation of tagmentation-based NGS library preparation. An example of the power of tagmentation was demonstrated in the transition from CIRCLE-seq to CHANGE-seq1. While CIRCLE-seq provided a sensitive, unbiased biochemical method for detecting off-target cleavage, the original protocol relied on more labor-intensive library preparation. The advancement to CHANGE-seq integrated tagmentation, dramatically simplifying the workflow. By simultaneously fragmenting and tagging DNA with sequencing adapters, tagmentation enables faster, more efficient library preparation, reducing bottlenecks and improving reproducibility. This shift sets the stage for further innovation across other off-target assays like GUIDE-seq (Genome-wide, Unbiased Identification of Double-Stranded Breaks Enabled by Sequencing).
The GUIDE-seq Challenge
GUIDE-seq has been a cornerstone assay for cellular measurement of CRISPR off-target activity since it was first published in 2015 by Tsai, et.al. in Nature Biotechnology2. By leveraging double-stranded oligodeoxynucleotide (dsODN) integration at double-stranded breaks, GUIDE-seq provides powerful insights into real-world off-target events within cellular contexts. However, the method suffers from limitations:
- Complex library preparation: Traditional workflows require multiple enzymatic steps, ligations, and nested PCR, increasing time and variability. Library preparation takes an 8-hour day to complete (Figure 1).
- Lower throughput: The hands-on nature limits scalability, constraining adoption for large-scale studies.
- Sensitivity trade-offs: The complex workflow can reduce sensitivity and reproducibility, particularly in challenging samples.
These limitations leave a gap between GUIDE-seq’s powerful biology and the streamlined, NGS-compatible workflows demanded in today’s gene editing landscape.
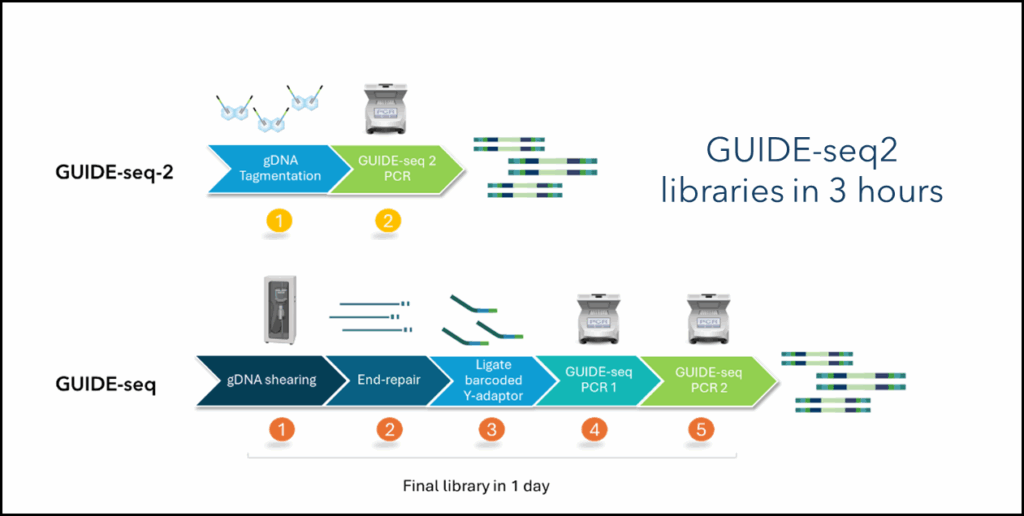
Figure 1. GUIDE-seq-23 workflow compared to the original GUIDE-seq protocol4. Tagmentation saves several steps significantly contributing to the reduction in library preparation time.
Enter GUIDE-seq2: Power Meets Efficiency
The introduction of GUIDE-seq2 in 2025 marks a major milestone in addressing these challenges. By integrating tagmentation into the library preparation workflow, GUIDE-seq2 delivers a more sensitive, reproducible and scalable solution while retaining the assay’s proven biology. Second-generation GUIDE-seq-2 has been validated across five therapeutic loci in human primary T cells, demonstrating strong correlation with the original GUIDE-seq method, establishing the ability to evolve the assay without compromising assay results3.
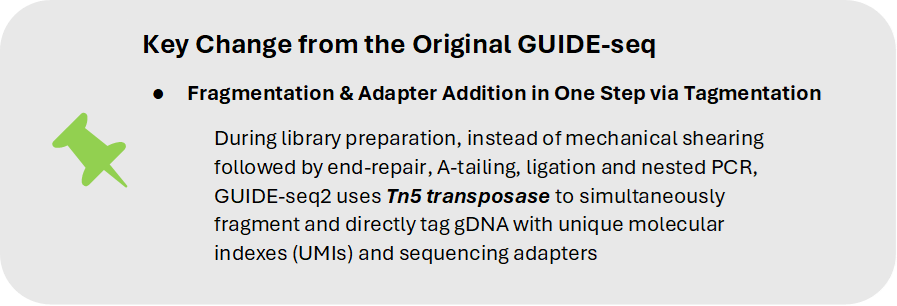
The original GUIDE-seq library preparation method requires fragmentation of gDNA (often using mechanical or enzymatic shearing) followed by end-repair/A-tailing of resulting fragments, ligation and nested PCR of library molecules. GUIDE-seq-2 replaces physical DNA shearing and multiple enzymatic and cleanup steps with tagmentation, while also eliminating the need for nested PCR. Tn5 transposase is loaded with a unique molecular index (UMI) and i5 adaptor (commercially available as Tagify from seqWell) which is directly tagged onto DNA during enzyme fragmentation. Tagmented gDNA undergoes a single round of PCR amplification using a tag-specific primer with an i7 barcode primer.
“The simplified GUIDE-seq-2 workflow substantially streamlines the process and enables high-throughput experiments, while also decreasing the requirement of input genomic DNA for library preparation by approximately 4-fold.” – Developers of GUIDE-seq23
GUIDE-seq2 advantages include:
- Streamlined workflow: Tagmentation eliminates multiple library prep steps, reducing assay complexity and hands-on time. Total library preparation can now be completed in 3 hours.
- Scalability: GUIDE-seq2 can be readily scaled for large panels or therapeutic pipelines, meeting the needs of high-throughput labs.
- Improved reproducibility: Simplified workflows reduce sources of technical variability, making results more consistent across experiments.
- Compatibility with modern sequencing: GUIDE-seq2 aligns with the latest NGS platforms and best practices, ensuring smoother integration into existing workflows.
Population-Scale Off-Target Effects
As genome editing technologies such as CRISPR-Cas9 move closer to population and clinical applications, it has become clear that genome-based off-target analysis using reference genomes alone is not enough. Even small genetic variants—such as single nucleotide polymorphisms (SNPs) or structural variations—can create or abolish CRISPR off-target sites, substantially altering editing specificity. This means that assays limited to a single genome background risk overlooking clinically relevant off-targets in diverse populations. Incorporating population-scale off-target analysis ensures that editing outcomes are evaluated across the spectrum of human genetic diversity, providing greater confidence in safety, sensitivity, and therapeutic applicability. The evolution to GUIDE-seq2 now offers an off-target assay that fulfills the throughput requirement and commercial availability of critical assay reagents to power large-scale studies.
GUIDE-seq2 Publication SNIP-IT: Population-scale cellular GUIDE-seq2 and biochemical CHANGE-seq-R profiles reveal human genetic variation frequently affects Cas9 off-target activity.
Lazzarotto et al. developed GUIDE-seq-2 to better understand how genetic variation influences Cas9 off-target activity in human cells. Using the new method, they analyzed 665 GUIDE-seq2 libraries from six gRNA targets in lymphoblastoid cells across 95 individuals representing four ethnic groups:
- African ancestry (Southwest U.S.) – ASW
- European ancestry (Utah) – CEU
- Han Chinese (Beijing) – CHB
- Mexican ancestry (Los Angeles) – MXL
The results of their studies revealed that GUIDE-seq2 detected off-target events frequently overlapped with human genetic variants, the frequency of which depended on ethnicity. The highest frequency of overlap with one or more genetic variations was seen in the African ancestry group (16.6%) and the lowest in the Han Chinese group (7.8%). These findings indicate that individual genetic variants may frequently have unintended off-target activity and emphasize the importance of population-scale CRISPR analyses.
Looking Forward
The evolution from GUIDE-seq to GUIDE-seq2 mirrors the successful transition seen in CIRCLE-seq to CHANGE-seq. Tagmentation is redefining how off-target assays keep pace with the scale and precision of modern sequencing. For researchers and developers, GUIDE-seq2 represents not just an incremental update, but a transformational one—making in-cell off-target detection more accessible, reliable, and ready for population-scale studies.
In a landscape where regulatory scrutiny and clinical demands are rising, having assays that combine biological relevance with operational efficiency is critical. GUIDE-seq2 demonstrates how thoughtful technical innovation can unlock the next phase of genome editing research and development.
Enhancing Gene Editing QC with Tagify Adapter-Loaded Transposases
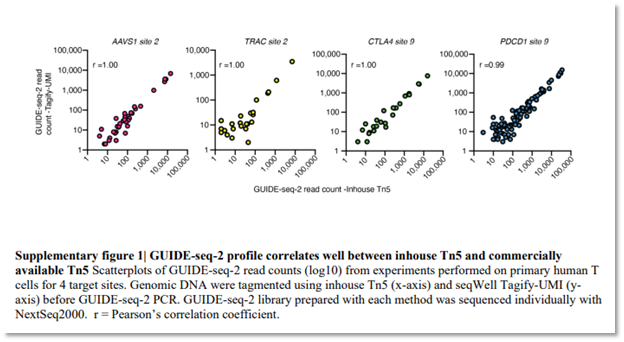
The GUIDE-seq-2 protocol incorporates Tn5 transposase loaded directly with P5/i5 sequence adapters and UMI indexes (Tagify™ i5 UMI from seqWell), demonstrating how loaded transposases can streamline workflows, increase efficiency, and reduce manual steps. While laboratories can express, purify and load Tn5 inhouse, the correlation to the commercially available counterpart provides researchers easy access to a highly consistent and fully-QC’d source of this critical assay reagent.
In the GUIDE-seq2 publication from Lazzarotto, et. al., the authors compared seqWell off-the-shelf Tagify™ i5 UMI reagent with lab-generated loaded transposase. They concluded that:
- Tagify i5 UMI provides a reliable, commercial source of loaded transposase that produces equivalent assay performance
- 99 – 1.00 correlation between seqWell’s Tagify and inhouse produced Tn5 for all four of the targets examined
Figure source: Lazzarotto, C, et.al., bioRxiv, 2025-02.
Evolve Your Assay Using Tagify™ Custom-loaded Transposases
In addition to Tagify i5 UMI, seqWell offers Tagify™ Custom-loaded Transposases: Tn5 or TnX transposase – our next-generation with enhanced sequencing performance – loaded with custom payloads to help drive other transposase-based assays, such as UDiTaS™, ATAC-Seq, SHARE-Seq, GUIDE-tag, CHANGE-Seq, TTIS-Seq, or RGen-Seq.
Contact us to learn how to convert your assay library preparation to a highly scalable tagmentation-based method using seqWell Tagify™ reagents
Read our Additional Blogs on Gene Editing QC
- Gaining Confidence in Gene Editing Analysis
- A Guide to Selecting the Right Gene Editing Off-Target Assay
References
- Lazzarotto, CR, Malinin, NL, Li, Y, Zhang, R, Yang, Y, Lee, G, Cowley, E, He, Y, Lan, X, Jividen, K, Katta, V, Kolmakova, NG, Petersen, CT, Qi, Q, Strelcov, E, Maragh, S, Krenciute, G, Ma, J, Cheng, Y, & Tsai, S. Q. (2020). CHANGE-seq reveals genetic and epigenetic effects on CRISPR-Cas9 genome-wide activity. Nat Biotechnol, 38(11), 1317-1327.
- Tsai, S. Q., Zheng, Z., Nguyen, N. T., Liebers, M., Topkar, V. V., Thapar, V., Wyvekens, N., Khayter, C., Le, L.P., Aryee, M. & Joung, J. K. (2015). GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol, 33(2), 187–197.
- Lazzarotto, C., Li, Y., Flory, A. R., Chyr, J., Yang, M., Katta V., Urbina, E., Lee, G., Wood, R., Matsubara, A., Rashkin, S., Ma, J., Cheng, Y., & Tsai, S.Q. (2025). Population-scale cellular GUIDE-seq-2 and biochemical CHANGE-seq-R profiles reveal human genetic variation frequently affects Cas9 off-target activity. bioRxiv, 2025-02.
- Malinin, N. L., Lee, G., Lazzarotto, C. R., Li, Y., Zheng, Z., Nguyen, N. T., Liebers, M., Topkar, V.V., Iafrate, A.J., Le, L.P., Aryee, M.J., Joung, J.K., … & Tsai, S. Q. (2021). Defining genome-wide CRISPR–Cas genome-editing nuclease activity with GUIDE-seq. Nat Protoc, 16(12), 5592-5615.