AgriPrep™ Library Prep Kit
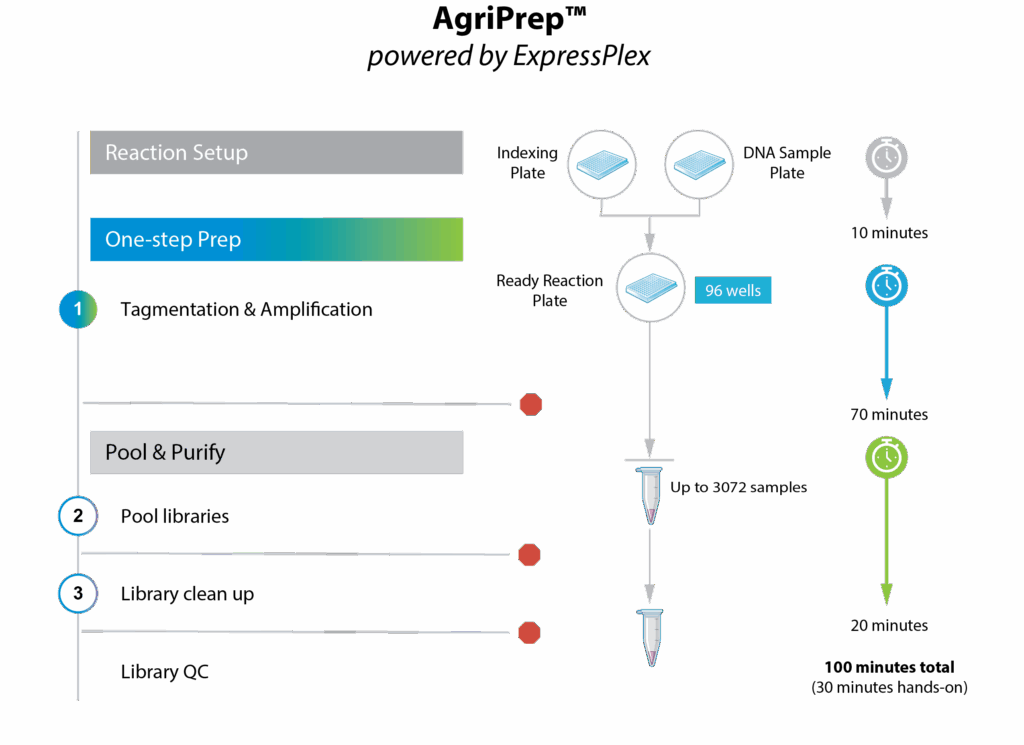
Bred for NGS library prep workflow simplicity and throughput! Powered by the one-step ExpressPlex workflow, AgriPrep is optimized for plant & animal low-pass WGS & SKIM-sequencing and goes from extracted genomic DNA to sequence-ready libraries in 100 minutes.
- Auto-normalization & early pooling
- Up to 3072 index combinations for scalability
- Easily miniaturized
- Significantly lower labor requirements
- Supports sustainability – up to 95% fewer consumables
>> PRESS RELEASE: Announcing Launch of AgriPrep Library Prep Kit
AgriPrep – Harvest actionable insights using simple, scalable, cost-effective sequence exploration!
Agrigenomics is increasingly relying on low-coverage whole genome sequencing (lcWGS). However, many NGS library prep methods fall short when scaled to the population level.
AgriPrep supports affordable, ultra-high-throughput plant & animal low-pass WGS & SKIM-sequencing approaches without sacrificing quality through unmatched protocol simplicity, auto-normalization, massive multiplexing, and lower all-in sequencing costs. AgriPrep is available in bulk reagent formats and can be performed in ultra-high-throughput formats including 384-well or Echo-compatible plates.
>> Simple, 100-minute workflow
>> Benefits of auto-normalization

Simple workflow
![]()
One-step tagmentation & amplification
Auto-normalization
Early sample pooling
100-minute workflow, 30 minutes hands-on
SCALABLE
![]()
Automation-friendly
Up to 3072 indexes
384-well plate format compatibility
Available in bulk reagent format
Affordable
![]()
Up to 95% reduction in consumables
Significant labor savings
Easily miniaturized

Scalable Genotyping By Sequencing (GBS) Using Low-Pass Whole Genome Sequencing (lpWGS) and a One-Step, TnX TransposaseBased Library Prep Method for Genomic Selection
Bred for NGS workflow simplicity and throughput
Powered by the 100-minute ExpressPlex Workflow
One-step tagmentation & amplification, auto-normalization, and early pooling come together creating the simplest library prep workflow on the market. Don’t let library prep be a bottleneck!
Auto-normalization maintains quality & provides cost efficiencies
Auto-normalization of mean insert size and read depth eliminates the need to individually quantify and dilute samples to ensure uniform coverage for all your critical samples.
Additional benefits include:
- Rapid protocol for higher laboratory throughput
- Labor and consumables cost savings
- Lower sequencing costs to meet coverage requirements for all samples
- Fewer opportunities for contamination or errors
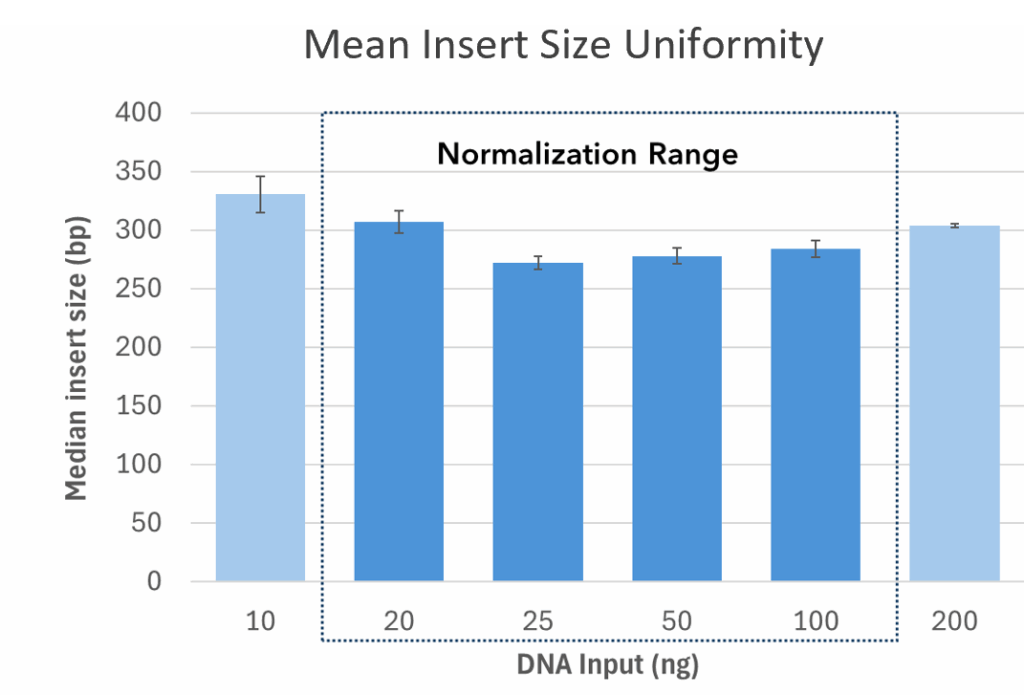
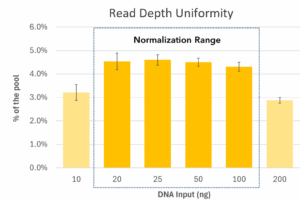
A 24-plex library pool was generated from 10 – 200 ng of Barley genomic DNA and sequenced on an Illumina MiSeq™ i100 system. AgriPrep has a working range of 10 to 200 ng of DNA input, with auto-normalization of both mean insert size and read depth from 20 to 100 ng. Mean insert size across the total working range (CV = 8.6%) and normalization range (CV = 6.9%). Read depth across the total working range (CV = 17.5%) and normalization range (CV = 10.5%)
Save Up to 95% on Consumables
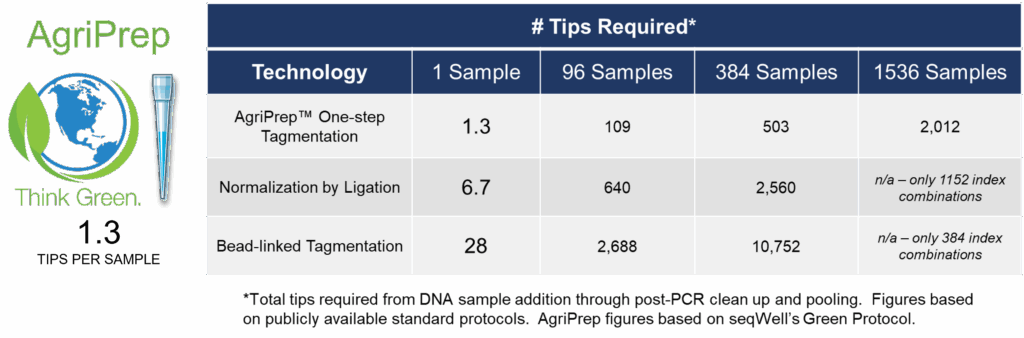
Streamlined Protocol & 3072 Indexes = Scalability, Sustainability & Affordability
AgriPrep’s pre-plated reagents, one-step tagmentation & amplification method, early sample pooling, and sequential reagent addition provide a highly streamlined workflow that results in the use of <2 tips/sample. When combined with the availability of 3072 indexes, AgriPrep offers unmatched scalability while minimizing environmental impact.
Lower sequencing costs through consistent coverage
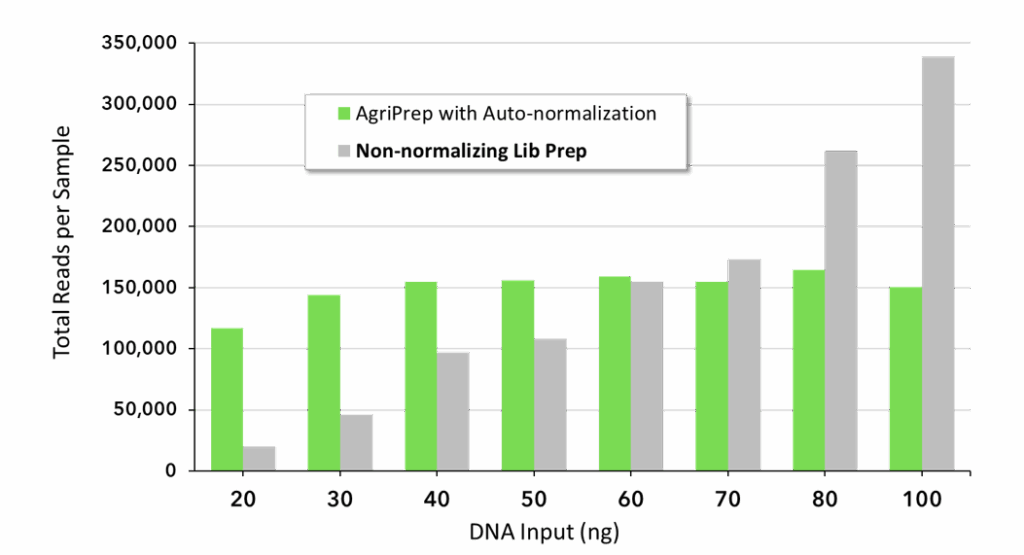
Don’t put your genotyping insights at risk due to the cost of over-sequencing. AgriPrep’s auto-normalization of read depth not only eliminates the costly time and labor requirements of quantifying and diluting individual samples, but importantly it minimizes differences in sequence coverage between samples. Thus, the need to over-sequence to attain the desired coverage levels of your critical samples can be significantly reduced.
Reduce required sequencing by 80%! AgriPrep with auto-normalization and a non-normalizing library prep method were each used to prepare 8 samples with variable starting DNA inputs between 20 – 100 ng. Following library prep, samples were pooled by equal volume per method for sequencing. If each pool was sequenced to 1.2 million reads and the target was ≥100,000 reads per sample, the AgriPrep pool with highly consistent read counts can achieve ≥100,000 reads for all samples during the sequencing run. However, the non-normalizing prep pool would require 5X more additional sequencing (6 million reads total) to bring the lowest represented sample in the pool to ≥100,000 reads due the high variance in read counts.

Recover More Data From Your Sequencing
Simply spike-in PhiRx™ to increase your index color-balancing!
PhiRx Indexed Control NOW included with every AgriPrep Library Prep Kit
Are you in need of bulk-dispensed product for your miniaturized reactions?
AgriPrep Specifications
| Primary Applications | Genotyping-by-sequencing (GBS), Low pass sequencing, SKIM-seq |
| Sample Input types | Plant and animal genomic DNA |
| Transposase | TnX – Next generation engineered transposase |
| Kit format |
|
| DNA Input | Working range: 10-200 ng
Normalization range: 20-100 ng |
| Total Library Prep Time | 100 minutes (30 minutes hands-on time) |
| Indexing Method | Combinatorial Dual Indexing |
| Number of Unique Index Combinations | Up to 1536 (off-the-shelf); up to 3072 via custom ordering |
| Batch Size | 8-96 samples; bulk reagents support 384-well & Echo-compatible plate formats |
AgriPrep Library Prep Kit Includes:
- Indexing Reaction Plate(s)
- Ready Reaction Mix Plate(s)
- MAGwise Paramagnetic Beads
- PhiRx Indexed Control
- Users do not need to supply polymerase, primers, or barcodes. These are contained within the supplied ready reaction mix.