Scalable library prep for low-pass whole genome sequencing
Low-pass whole-genome sequencing is an effective technical alternative to microarray-based genotyping.
The economics of using sequencing in place of microarrays is further increased by plexWell’s ability to create balanced pools of libraries that efficiently multiplex as many genomic samples as possible on high-output sequencing runs. seqWell has demonstrated the use of plexWell™ for routine low-pass human WGS applications, where we show robust and scalable genotype imputation accuracy in combination with Gencove’s imputation SaaS.
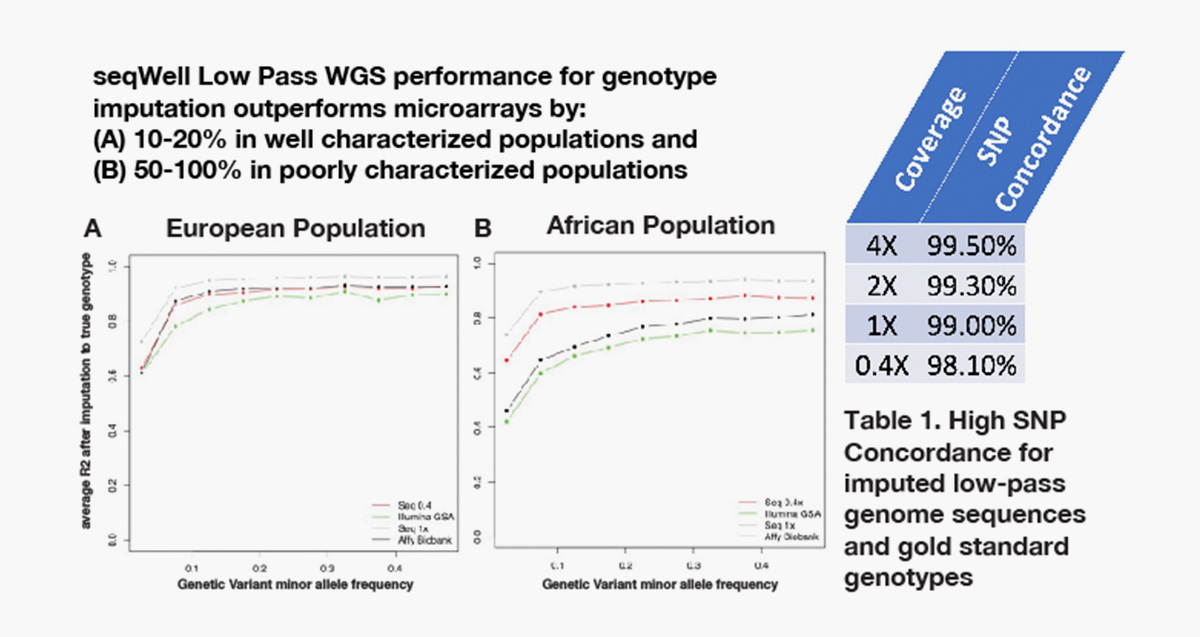
Gencove’s imputation SaaS in combination with seqWell’s low pass library preparation kit outperforms genotyping arrays.
“We have a 95-99% success rate with seqWell.”
Mohammad Faghihi. M.D., Ph.D., Sr. Vice President, Lab Operations, Galatea Bio
Mohammad Faghihi. M.D., Ph.D., is Sr. Vice President, Lab Operations, at Galatea Bio – a biotech company headquartered in Hialeah, Florida, made up of a global group of scientists working on filling the knowledge gap in population studies and its effect on underrepresented communities.
Here’s what he has to say about our plexWell LP384 library prep kit:
“We are doing low-pass whole genome sequencing (LP-WGS) to impute genotyping for Ancestry guidance and for making efficient genomic data available. We have tried LP-WGS products before, but seqWell’s was both faster and easier. Very early in the seqWell protocol, all samples get their unique indices and become combined into 1-2 tubes for remaining processing. The hands-on for index and further processing is short and generally requires less than 3 hours to complete.
Unlike full-day involved protocols from other companies, the simple seqWell workflow allows more throughput, even without using expensive robots. seqWell’s workflow also increases accuracy by providing unique sample indexing at the beginning without increased chance of index hopping.
Additionally, the number of failed samples is much lower, which saves time, money, and potential mislabelling caused by repeating experiments over and over. We were seeing a 75% success rate previously, but observed huge improvements. We now have a 95-99% success rate with seqWell.”
Low-pass WGS Resources
Robust, Scalable, Low-Coverage Whole Genome Sequencing for High-Throughput Crop Genotyping
In this poster we demonstrate the robust combination of the EchoLUTION Plant DNA extraction kit (BioEcho Life Sciences) and plexWell™ Low-Pass 384 (LP384) Library Preparation kit (seqWell) across multiple plant species with a range of genome sizes. The combined workflow features normalized multiplexed libraries without the need for time-consuming individual adjustment of input DNA, significantly simplifying the complex task of high-level multiplexing and enabling faster, cost-effective, and reliable results for routine crop genotyping applications such as principal component analysis.
Imputation of Loss-of-Function Genotypes Using Low Coverage WGS in a Cattle Population
Low-coverage whole genome sequencing (lcWGS) is an increasingly attractive alternative to microarray genotyping, as the cost of sequencing data has dropped over the last decade. A key need for lcWGS is being able to generate multiplexed NGS libraries from genomic DNA in which individual sample libraries are sufficiently normalized to produce adequate coverage of each genome for accurate genotype imputation. This study evaluates the use of the plexWell Low-Pass 384 Library Preparation kit (seqWell, Inc) for lcWGS of a cattle breeding population.
Low-Pass Whole-Genome Sequencing on the Element AVITI™ System Enables Cost-Effective Genotyping
Low-pass whole-genome sequencing enabled by scalable library prep offers a competitive alternative to microarray-based genotyping
plexWell Low Pass 384 Library Preparation Kit: Cost-effective high-throughput low pass genotyping
High-throughput library preparation for low-pass sequence-based genotyping pipelines
seqWell+Gencove Press Release
Looking for more information?
For more information about our scalable workflow solutions for low-pass whole genome sequencing, please contact us to work with one of our product specialists.
